UniProt: the universal protein knowledgebase
- PMID: 27899622
- PMCID: PMC5210571
- DOI: 10.1093/nar/gkw1099
UniProt: the universal protein knowledgebase
Erratum in
-
UniProt: the universal protein knowledgebase.Nucleic Acids Res. 2018 Mar 16;46(5):2699. doi: 10.1093/nar/gky092. Nucleic Acids Res. 2018. PMID: 29425356 Free PMC article. No abstract available.
Abstract
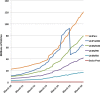
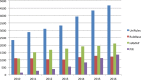
The UniProt knowledgebase is a large resource of protein sequences and associated detailed annotation. The database contains over 60 million sequences, of which over half a million sequences have been curated by experts who critically review experimental and predicted data for each protein. The remainder are automatically annotated based on rule systems that rely on the expert curated knowledge. Since our last update in 2014, we have more than doubled the number of reference proteomes to 5631, giving a greater coverage of taxonomic diversity. We implemented a pipeline to remove redundant highly similar proteomes that were causing excessive redundancy in UniProt. The initial run of this pipeline reduced the number of sequences in UniProt by 47 million. For our users interested in the accessory proteomes, we have made available sets of pan proteome sequences that cover the diversity of sequences for each species that is found in its strains and sub-strains. To help interpretation of genomic variants, we provide tracks of detailed protein information for the major genome browsers. We provide a SPARQL endpoint that allows complex queries of the more than 22 billion triples of data in UniProt (http://sparql.uniprot.org/). UniProt resources can be accessed via the website at http://www.uniprot.org/.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
UniProt: the universal protein knowledgebase in 2021.Nucleic Acids Res. 2021 Jan 8;49(D1):D480-D489. doi: 10.1093/nar/gkaa1100. Nucleic Acids Res. 2021. PMID: 33237286 Free PMC article.
-
UniProt: the Universal Protein knowledgebase.Nucleic Acids Res. 2004 Jan 1;32(Database issue):D115-9. doi: 10.1093/nar/gkh131. Nucleic Acids Res. 2004. PMID: 14681372 Free PMC article.
-
UniProtKB/Swiss-Prot, the Manually Annotated Section of the UniProt KnowledgeBase: How to Use the Entry View.Methods Mol Biol. 2016;1374:23-54. doi: 10.1007/978-1-4939-3167-5_2. Methods Mol Biol. 2016. PMID: 26519399
-
In silico characterization of proteins: UniProt, InterPro and Integr8.Mol Biotechnol. 2008 Feb;38(2):165-77. doi: 10.1007/s12033-007-9003-x. Epub 2007 Oct 4. Mol Biotechnol. 2008. PMID: 18219596 Review.
-
Bioinformatics Tools for Proteomics Data Interpretation.Adv Exp Med Biol. 2016;919:281-341. doi: 10.1007/978-3-319-41448-5_16. Adv Exp Med Biol. 2016. PMID: 27975225 Review.
Cited by
-
Molecular mechanisms of sulforaphane in Alzheimer's disease: insights from an in-silico study.In Silico Pharmacol. 2024 Nov 1;12(2):96. doi: 10.1007/s40203-024-00267-4. eCollection 2024. In Silico Pharmacol. 2024. PMID: 39493676 Free PMC article.
-
The proteome of the human endolymphatic sac endolymph.Sci Rep. 2021 Jun 4;11(1):11850. doi: 10.1038/s41598-021-89597-3. Sci Rep. 2021. PMID: 34088924 Free PMC article.
-
A putative novel starch-binding domain revealed by in silico analysis of the N-terminal domain in bacterial amylomaltases from the family GH77.3 Biotech. 2021 May;11(5):229. doi: 10.1007/s13205-021-02787-8. Epub 2021 Apr 21. 3 Biotech. 2021. PMID: 33968573 Free PMC article.
-
Molecular and cellular paradigms of multidrug resistance in cancer.Cancer Rep (Hoboken). 2022 Dec;5(12):e1291. doi: 10.1002/cnr2.1291. Epub 2020 Oct 13. Cancer Rep (Hoboken). 2022. PMID: 33052041 Free PMC article. Review.
-
Comparative Genomics of Peroxisome Biogenesis Proteins: Making Sense of the PEX Proteins.Front Cell Dev Biol. 2021 May 20;9:654163. doi: 10.3389/fcell.2021.654163. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34095119 Free PMC article.
References
-
- Suzek B.E., Huang H., McGarvey P., Mazumder R., Wu C.H.. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics. 2007; 23:1282–1288. - PubMed
-
- Leinonen R., Diez F.G., Binns D., Fleischmann W., Lopez R., Apweiler R.. UniProt archive. Bioinformatics. 2004; 20:3236–3237. - PubMed
-
- Giraldo-Calderon G.I., Emrich S.J., MacCallum R.M., Maslen G., Dialynas E., Topalis P., Ho N., Gesing S., VectorBase C., Madey G. et al. . VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 2015; 43:D707–D713. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

