Exploring the impact of EGFR T790M neighboring SNPs on ARMS-based T790M mutation assay
- PMID: 27895798
- PMCID: PMC5104263
- DOI: 10.3892/ol.2016.5184
Exploring the impact of EGFR T790M neighboring SNPs on ARMS-based T790M mutation assay
Abstract
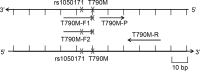
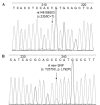
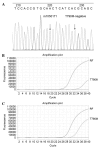
The present study aimed to explore the influence of T790M neighboring single nucleotide polymorphism (SNP) on the sensitivity of amplification refractory mutation system (ARMS)-based T790M mutation assay. Three ARMS-quantitative polymerase chain reaction (qPCR) systems (system 1 had a forward ARMS primer without rs1050171, system 2 included a forward ARMS primer with rs1050171 and system 3 contained the above two forward ARMS primers) were used to detect the T790M mutation in two series plasmid samples and genomic DNA (gDNA) of the cell line H1975. A total of 670 formalin-fixed paraffin-embedded (FFPE) tumor samples from non-small cell lung cancer patients were used to detect the epidermal growth factor receptor (EGFR) gene T790M mutation by direct sequencing and ARMS-qPCR. The ARMS-qPCR system 1 effectively detected samples with as low as 1% T790M mutant plasmid 1 (without rs1050171) and with 50% T790M mutant plasmid 2 (with rs1050171), while the ARMS-qPCR system 2 detected samples with 20 and 50% T790M mutant plasmid 1, in addition to samples with 1% T790M mutant plasmid 2. For the ARMS-qPCR system 3, samples with as low as 1% T790M mutant plasmids 1 or 2 were effectively detected. For gDNA analysis of the cell line H1975, the T790M mutation was effectively detected by the ARMS-qPCR systems 2 and 3 (~50% mutation rate), but was detected with a low mutation abundance by the ARMS-qPCR system 1 (~1% mutation rate). Of the 670 FFPE samples, 5 cases were identified to have the T790M mutation by sequencing and by the ARMS-qPCR system 1. One sample (named N067), which was considered as T790M-negative by sequencing, was demonstrated to have the T790M mutation using the ARMS-qPCR system 1. Sample N094, which was variant homozygous for rs1050171 and was indicated to be T790M-negative by sequencing and by the ARMS-qPCR system 1, was identified to have the T790M mutation with the ARMS-qPCR system 3. The A-variant allele frequency of rs1050171 was observed to be 28.2% in the 670 FFPE tumor samples, while the presence of rs148188503 (c. C2355T, p. T785T) was observed in sample N558, and a novel SNP with a base substitution (c. T2375C) at position 792 (p. L792P) in exon 20 of the EGFR gene was observed in sample N310. rs1050171 is a high-frequency SNP located near T790M, and the mutation statuses of rs1050171 appear to influence the sensitivity of the ARMS-based T790M detection system, thus generating a 14.3% false-negative rate (1/7). The present study proposes the risk that target neighboring SNPs (as far as 8 bp away in the present study) may exert on the sensitivity of ARMS-based detection methods.
Keywords: amplification refractory mutation system; epidermal growth factor receptor gene T790M mutation; non-small cell lung cancer; rs1050171; single nucleotide polymorphism.
Figures

Similar articles
-
Comparison of droplet digital PCR and conventional quantitative PCR for measuring EGFR gene mutation.Exp Ther Med. 2015 Apr;9(4):1383-1388. doi: 10.3892/etm.2015.2221. Epub 2015 Jan 27. Exp Ther Med. 2015. PMID: 25780439 Free PMC article.
-
Highly sensitive and noninvasive detection of epidermal growth factor receptor T790M mutation in non-small cell lung cancer.Clin Chim Acta. 2013 Oct 21;425:119-24. doi: 10.1016/j.cca.2013.07.012. Epub 2013 Jul 23. Clin Chim Acta. 2013. PMID: 23886554
-
EGFR T790M detection in formalin-fixed paraffin-embedded tissues of patients with lung cancer using RNA-based in situ hybridization: A preliminary feasibility study.Thorac Cancer. 2019 Oct;10(10):1936-1944. doi: 10.1111/1759-7714.13169. Epub 2019 Aug 13. Thorac Cancer. 2019. PMID: 31407509 Free PMC article.
-
[Evaluation of Consistency in detection of epidermal growth factor receptor gene T790M mutation in plasma and tumor specimens of patients with lung adenocarcinoma].Zhonghua Zhong Liu Za Zhi. 2018 Jan 23;40(1):35-39. doi: 10.3760/cma.j.issn.0253-3766.2018.01.006. Zhonghua Zhong Liu Za Zhi. 2018. PMID: 29365415 Chinese.
-
A sensitive and practical method to detect the T790M mutation in the epidermal growth factor receptor.Oncol Lett. 2016 Apr;11(4):2573-2579. doi: 10.3892/ol.2016.4263. Epub 2016 Feb 23. Oncol Lett. 2016. PMID: 27073519 Free PMC article.
Cited by
-
A comparison of MASS-PCR and ARMS-PCR for the detection of lung cancer gene mutation.Transl Cancer Res. 2019 Nov;8(7):2564-2569. doi: 10.21037/tcr.2019.10.37. Transl Cancer Res. 2019. PMID: 35117013 Free PMC article.
-
Lung cancer mutation testing: a clinical retesting study of agreement between a real-time PCR and a mass spectrometry test.Oncotarget. 2017 Sep 16;8(60):101437-101451. doi: 10.18632/oncotarget.21023. eCollection 2017 Nov 24. Oncotarget. 2017. PMID: 29254176 Free PMC article.
-
Two missense variants of the epidermal growth factor receptor gene are associated with non small cell lung carcinoma in the subjects from Iraq.Mol Biol Rep. 2022 Dec;49(12):11653-11661. doi: 10.1007/s11033-022-07933-w. Epub 2022 Sep 28. Mol Biol Rep. 2022. PMID: 36169894
-
Evaluation of PCR-HRM, RFLP, and direct sequencing as simple and cost-effective methods to detect common EGFR mutations in plasma cell-free DNA of non-small cell lung cancer patients.Cancer Rep (Hoboken). 2019 Aug;2(4):e1159. doi: 10.1002/cnr2.1159. Epub 2019 Feb 3. Cancer Rep (Hoboken). 2019. PMID: 32721094 Free PMC article.
-
Detection of somatic mutations in ctDNA derived from adenocarcinoma patients - EGFR tyrosine kinase inhibitor monitoring preliminary study.Contemp Oncol (Pozn). 2019;23(2):87-91. doi: 10.5114/wo.2019.85879. Epub 2019 Jun 13. Contemp Oncol (Pozn). 2019. PMID: 31316290 Free PMC article.
References
-
- Kuang Y, Rogers A, Yeap BY, Wang L, Makrigiorgos M, Vetrand K, Thiede S, Distel RJ, Jänne PA. Noninvasive detection of EGFR T790M in gefitinib or erlotinib resistant non-small cell lung cancer. Clinical cancer research: An official journal of the American Association for Cancer Research. 2009;15:2630–2636. doi: 10.1158/1078-0432.CCR-08-2592. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
