Structural basis for the recognition of spliceosomal SmN/B/B' proteins by the RBM5 OCRE domain in splicing regulation
- PMID: 27894420
- PMCID: PMC5127646
- DOI: 10.7554/eLife.14707
Structural basis for the recognition of spliceosomal SmN/B/B' proteins by the RBM5 OCRE domain in splicing regulation
Abstract
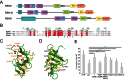
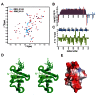
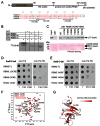
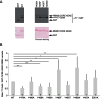
The multi-domain splicing factor RBM5 regulates the balance between antagonistic isoforms of the apoptosis-control genes FAS/CD95, Caspase-2 and AID. An OCRE (OCtamer REpeat of aromatic residues) domain found in RBM5 is important for alternative splicing regulation and mediates interactions with components of the U4/U6.U5 tri-snRNP. We show that the RBM5 OCRE domain adopts a unique β-sheet fold. NMR and biochemical experiments demonstrate that the OCRE domain directly binds to the proline-rich C-terminal tail of the essential snRNP core proteins SmN/B/B'. The NMR structure of an OCRE-SmN peptide complex reveals a specific recognition of poly-proline helical motifs in SmN/B/B'. Mutation of conserved aromatic residues impairs binding to the Sm proteins in vitro and compromises RBM5-mediated alternative splicing regulation of FAS/CD95. Thus, RBM5 OCRE represents a poly-proline recognition domain that mediates critical interactions with the C-terminal tail of the spliceosomal SmN/B/B' proteins in FAS/CD95 alternative splicing regulation.
Keywords: NMR-spectroscopy; OCRE domain; alternative splicing; biochemistry; biophysics; human; poly-proline; protein-protein interactions; structural biology.
Conflict of interest statement
JV: Reviewing editor, eLife. The other authors declare that no competing interests exist.
Figures



Similar articles
-
RBM5/Luca-15/H37 regulates Fas alternative splice site pairing after exon definition.Mol Cell. 2008 Oct 10;32(1):81-95. doi: 10.1016/j.molcel.2008.08.008. Mol Cell. 2008. PMID: 18851835
-
Nuclear Magnetic Resonance Structure of a Novel Globular Domain in RBM10 Containing OCRE, the Octamer Repeat Sequence Motif.Structure. 2016 Jan 5;24(1):158-164. doi: 10.1016/j.str.2015.10.029. Epub 2015 Dec 17. Structure. 2016. PMID: 26712279 Free PMC article.
-
Structural basis for specific RNA recognition by the alternative splicing factor RBM5.Nat Commun. 2023 Jul 15;14(1):4233. doi: 10.1038/s41467-023-39961-w. Nat Commun. 2023. PMID: 37454201 Free PMC article.
-
The SMN complex: an assembly machine for RNPs.Cold Spring Harb Symp Quant Biol. 2006;71:313-20. doi: 10.1101/sqb.2006.71.001. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381311 Review.
-
Structural dynamics of the N-terminal domain and the Switch loop of Prp8 during spliceosome assembly and activation.Nucleic Acids Res. 2018 May 4;46(8):3833-3840. doi: 10.1093/nar/gky242. Nucleic Acids Res. 2018. PMID: 29635373 Free PMC article. Review.
Cited by
-
Neurochondrin interacts with the SMN protein suggesting a novel mechanism for spinal muscular atrophy pathology.J Cell Sci. 2018 Apr 17;131(8):jcs211482. doi: 10.1242/jcs.211482. J Cell Sci. 2018. PMID: 29507115 Free PMC article.
-
Splicing Site Recognition by Synergy of Three Domains in Splicing Factor RBM10.Biochemistry. 2018 Mar 13;57(10):1563-1567. doi: 10.1021/acs.biochem.7b01242. Epub 2018 Feb 16. Biochemistry. 2018. PMID: 29450990 Free PMC article.
-
Clinical diagnostic exome evaluation for an infant with a lethal disorder: genetic diagnosis of TARP syndrome and expansion of the phenotype in a patient with a newly reported RBM10 alteration.BMC Med Genet. 2017 Jun 2;18(1):60. doi: 10.1186/s12881-017-0426-3. BMC Med Genet. 2017. PMID: 28577551 Free PMC article.
-
RNA Binding Motif 5 (RBM5) in the CNS-Moving Beyond Cancer to Harness RNA Splicing to Mitigate the Consequences of Brain Injury.Front Mol Neurosci. 2020 Jul 15;13:126. doi: 10.3389/fnmol.2020.00126. eCollection 2020. Front Mol Neurosci. 2020. PMID: 32765218 Free PMC article.
-
Yeast Spt6 Reads Multiple Phosphorylation Patterns of RNA Polymerase II C-Terminal Domain In Vitro.J Mol Biol. 2020 Jun 26;432(14):4092-4107. doi: 10.1016/j.jmb.2020.05.007. Epub 2020 May 19. J Mol Biol. 2020. PMID: 32439331 Free PMC article.
References
-
- Bacrot S, Doyard M, Huber C, Alibeu O, Feldhahn N, Lehalle D, Lacombe D, Marlin S, Nitschke P, Petit F, Vazquez MP, Munnich A, Cormier-Daire V. Mutations in SNRPB, encoding components of the core splicing machinery, cause cerebro-costo-mandibular syndrome. Human Mutation. 2015;36:187–190. doi: 10.1002/humu.22729. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

