The H3K9 dimethyltransferases EHMT1/2 protect against pathological cardiac hypertrophy
- PMID: 27893464
- PMCID: PMC5199699
- DOI: 10.1172/JCI88353
The H3K9 dimethyltransferases EHMT1/2 protect against pathological cardiac hypertrophy
Abstract
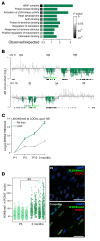
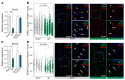
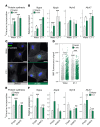
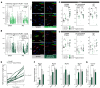
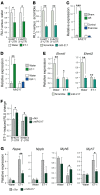
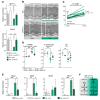
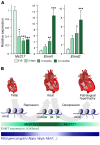
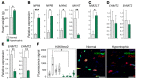
Cardiac hypertrophic growth in response to pathological cues is associated with reexpression of fetal genes and decreased cardiac function and is often a precursor to heart failure. In contrast, physiologically induced hypertrophy is adaptive, resulting in improved cardiac function. The processes that selectively induce these hypertrophic states are poorly understood. Here, we have profiled 2 repressive epigenetic marks, H3K9me2 and H3K27me3, which are involved in stable cellular differentiation, specifically in cardiomyocytes from physiologically and pathologically hypertrophied rat hearts, and correlated these marks with their associated transcriptomes. This analysis revealed the pervasive loss of euchromatic H3K9me2 as a conserved feature of pathological hypertrophy that was associated with reexpression of fetal genes. In hypertrophy, H3K9me2 was reduced following a miR-217-mediated decrease in expression of the H3K9 dimethyltransferases EHMT1 and EHMT2 (EHMT1/2). miR-217-mediated, genetic, or pharmacological inactivation of EHMT1/2 was sufficient to promote pathological hypertrophy and fetal gene reexpression, while suppression of this pathway protected against pathological hypertrophy both in vitro and in mice. Thus, we have established a conserved mechanism involving a departure of the cardiomyocyte epigenome from its adult cellular identity to a reprogrammed state that is accompanied by reexpression of fetal genes and pathological hypertrophy. These results suggest that targeting miR-217 and EHMT1/2 to prevent H3K9 methylation loss is a viable therapeutic approach for the treatment of heart disease.
Conflict of interest statement
W. Reik is a consultant and shareholder of Cambridge Epigenetix.
Figures


Similar articles
-
Epigenetic response to environmental stress: Assembly of BRG1-G9a/GLP-DNMT3 repressive chromatin complex on Myh6 promoter in pathologically stressed hearts.Biochim Biophys Acta. 2016 Jul;1863(7 Pt B):1772-81. doi: 10.1016/j.bbamcr.2016.03.002. Epub 2016 Mar 4. Biochim Biophys Acta. 2016. PMID: 26952936 Free PMC article.
-
Histone Methyltransferase G9a Is Required for Cardiomyocyte Homeostasis and Hypertrophy.Circulation. 2017 Sep 26;136(13):1233-1246. doi: 10.1161/CIRCULATIONAHA.117.028561. Epub 2017 Aug 4. Circulation. 2017. PMID: 28778944
-
EHMT1 and EHMT2 inhibition induces fetal hemoglobin expression.Blood. 2015 Oct 15;126(16):1930-9. doi: 10.1182/blood-2015-06-649087. Epub 2015 Aug 28. Blood. 2015. PMID: 26320100 Free PMC article.
-
Regulation of cell differentiation and function by the euchromatin histone methyltranserfases G9a and GLP.Biochem Cell Biol. 2016 Feb;94(1):26-32. doi: 10.1139/bcb-2015-0017. Epub 2015 Jun 1. Biochem Cell Biol. 2016. PMID: 26198080 Review.
-
MicroRNAs in a hypertrophic heart: from foetal life to adulthood.Biol Rev Camb Philos Soc. 2017 Aug;92(3):1314-1331. doi: 10.1111/brv.12283. Epub 2016 Jun 1. Biol Rev Camb Philos Soc. 2017. PMID: 27247253 Review.
Cited by
-
Plasma metabolomics profiles in Black and White participants of the Adventist Health Study-2 cohort.BMC Med. 2023 Oct 31;21(1):408. doi: 10.1186/s12916-023-03101-4. BMC Med. 2023. PMID: 37904137 Free PMC article.
-
Calcium Signaling in Cardiomyocyte Function.Cold Spring Harb Perspect Biol. 2020 Mar 2;12(3):a035428. doi: 10.1101/cshperspect.a035428. Cold Spring Harb Perspect Biol. 2020. PMID: 31308143 Free PMC article. Review.
-
Inhibition of the extracellular enzyme A disintegrin and metalloprotease with thrombospondin motif 4 prevents cardiac fibrosis and dysfunction.Cardiovasc Res. 2023 Aug 19;119(10):1915-1927. doi: 10.1093/cvr/cvad078. Cardiovasc Res. 2023. PMID: 37216909 Free PMC article.
-
Cardiomyocyte renewal in the human heart: insights from the fall-out.Eur Heart J. 2017 Aug 7;38(30):2333-2342. doi: 10.1093/eurheartj/ehx343. Eur Heart J. 2017. PMID: 28810672 Free PMC article. Review.
-
Genes encoding ACE2, TMPRSS2 and related proteins mediating SARS-CoV-2 viral entry are upregulated with age in human cardiomyocytes.J Mol Cell Cardiol. 2020 Oct;147:88-91. doi: 10.1016/j.yjmcc.2020.08.009. Epub 2020 Aug 18. J Mol Cell Cardiol. 2020. PMID: 32818486 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

