Structure-based design of broadly protective group a streptococcal M protein-based vaccines
- PMID: 27890396
- PMCID: PMC5143202
- DOI: 10.1016/j.vaccine.2016.11.065
Structure-based design of broadly protective group a streptococcal M protein-based vaccines
Abstract
Background: A major obstacle to the development of broadly protective M protein-based group A streptococcal (GAS) vaccines is the variability within the N-terminal epitopes that evoke potent bactericidal antibodies. The concept of M type-specific protective immune responses has recently been challenged based on the observation that multivalent M protein vaccines elicited cross-reactive bactericidal antibodies against a number of non-vaccine M types of GAS. Additionally, a new "cluster-based" typing system of 175M proteins identified a limited number of clusters containing closely related M proteins. In the current study, we used the emm cluster typing system, in combination with computational structure-based peptide modeling, as a novel approach to the design of potentially broadly protective M protein-based vaccines.
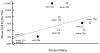
Methods: M protein sequences (AA 16-50) from the E4 cluster containing 17 emm types of GAS were analyzed using de novo 3-D structure prediction tools and the resulting structures subjected to chemical diversity analysis to identify sequences that were the most representative of the 3-D physicochemical properties of the M peptides in the cluster. Five peptides that spanned the range of physicochemical attributes of all 17 peptides were used to formulate synthetic and recombinant vaccines. Rabbit antisera were assayed for antibodies that cross-reacted with E4 peptides and whole bacteria by ELISA and for bactericidal activity against all E4GAS.
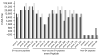
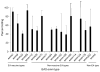
Results: The synthetic vaccine rabbit antisera reacted with all 17 E4M peptides and demonstrated bactericidal activity against 15/17 E4GAS. A recombinant hybrid vaccine containing the same E4 peptides also elicited antibodies that cross-reacted with all E4M peptides.
Conclusions: Comprehensive studies using structure-based design may result in a broadly protective M peptide vaccine that will elicit cluster-specific and emm type-specific antibody responses against the majority of clinically relevant emm types of GAS.
Keywords: Broadly neutralizing antibodies; Group A streptococcal vaccine; M protein; Structure-based design.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Conflict of interest statement
JBD is the inventor of certain technologies related to the development of group A streptococcal vaccines. The University of Tennessee Research Foundation has licensed the technology to Vaxent, LLC, of which JBD is the Chief Scientific Officer and a member. All other authors have no conflicts.
Figures

Similar articles
-
Bactericidal activity of M protein conserved region antibodies against group A streptococcal isolates from the Northern Thai population.BMC Microbiol. 2006 Aug 9;6:71. doi: 10.1186/1471-2180-6-71. BMC Microbiol. 2006. PMID: 16895610 Free PMC article.
-
Evaluation of novel Streptococcus pyogenes vaccine candidates incorporating multiple conserved sequences from the C-repeat region of the M-protein.Vaccine. 2012 Mar 9;30(12):2197-205. doi: 10.1016/j.vaccine.2011.12.115. Epub 2012 Jan 20. Vaccine. 2012. PMID: 22265945
-
New 30-valent M protein-based vaccine evokes cross-opsonic antibodies against non-vaccine serotypes of group A streptococci.Vaccine. 2011 Oct 26;29(46):8175-8. doi: 10.1016/j.vaccine.2011.09.005. Epub 2011 Sep 13. Vaccine. 2011. PMID: 21920403 Free PMC article.
-
Lipid core peptide technology and group A streptococcal vaccine delivery.Expert Rev Vaccines. 2004 Feb;3(1):43-58. doi: 10.1586/14760584.3.1.43. Expert Rev Vaccines. 2004. PMID: 14761243 Review.
-
Molecular biology of Group A Streptococcus and its implications in vaccine strategies.Indian J Med Microbiol. 2017 Apr-Jun;35(2):176-183. doi: 10.4103/ijmm.IJMM_17_16. Indian J Med Microbiol. 2017. PMID: 28681803 Review.
Cited by
-
Supercomputer-Based Ensemble Docking Drug Discovery Pipeline with Application to Covid-19.J Chem Inf Model. 2020 Dec 28;60(12):5832-5852. doi: 10.1021/acs.jcim.0c01010. Epub 2020 Dec 16. J Chem Inf Model. 2020. PMID: 33326239 Free PMC article.
-
Novel Big Data-Driven Machine Learning Models for Drug Discovery Application.Molecules. 2022 Jan 18;27(3):594. doi: 10.3390/molecules27030594. Molecules. 2022. PMID: 35163865 Free PMC article.
-
Variation, Indispensability, and Masking in the M protein.Trends Microbiol. 2018 Feb;26(2):132-144. doi: 10.1016/j.tim.2017.08.002. Epub 2017 Aug 31. Trends Microbiol. 2018. PMID: 28867148 Free PMC article. Review.
-
Progress in the Development of Structure-Based Vaccines.Methods Mol Biol. 2022;2412:15-33. doi: 10.1007/978-1-0716-1892-9_2. Methods Mol Biol. 2022. PMID: 34918239 Review.
-
Group A Streptococcus Infections: Their Mechanisms, Epidemiology, and Current Scope of Vaccines.Cureus. 2022 Dec 30;14(12):e33146. doi: 10.7759/cureus.33146. eCollection 2022 Dec. Cureus. 2022. PMID: 36721580 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

