The innate immune receptor Dectin-2 mediates the phagocytosis of cancer cells by Kupffer cells for the suppression of liver metastasis
- PMID: 27872290
- PMCID: PMC5150405
- DOI: 10.1073/pnas.1617903113
The innate immune receptor Dectin-2 mediates the phagocytosis of cancer cells by Kupffer cells for the suppression of liver metastasis
Abstract
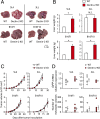
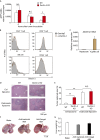
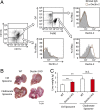
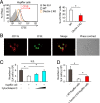
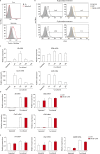
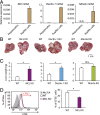
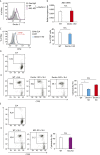
Tumor metastasis is the cause of most cancer deaths. Although metastases can form in multiple end organs, the liver is recognized as a highly permissive organ. Nevertheless, there is evidence for immune cell-mediated mechanisms that function to suppress liver metastasis by certain tumors, although the underlying mechanisms for the suppression of metastasis remain elusive. Here, we show that Dectin-2, a C-type lectin receptor (CLR) family of innate receptors, is critical for the suppression of liver metastasis of cancer cells. We provide evidence that Dectin-2 functions in resident macrophages in the liver, known as Kupffer cells, to mediate the uptake and clearance of cancer cells. Interestingly, Kupffer cells are selectively endowed with Dectin-2-dependent phagocytotic activity, with neither bone marrow-derived macrophages nor alveolar macrophages showing this potential. Concordantly, subcutaneous primary tumor growth and lung metastasis are not affected by the absence of Dectin-2. In addition, macrophage C-type lectin, a CLR known to be complex with Dectin-2, also contributes to the suppression of liver metastasis. Collectively, these results highlight the hitherto poorly understood mechanism of Kupffer cell-mediated control of metastasis that is mediated by the CLR innate receptor family, with implications for the development of anticancer therapy targeting CLRs.
Keywords: C-type lectin receptor; Dectin-2; Kupffer cell; liver metastasis; phagocytosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Opposite regulatory effects of IFN-β and IL-3 on C-type lectin receptors, antigen uptake, and phagocytosis in human macrophages.J Leukoc Biol. 2014 Jan;95(1):161-8. doi: 10.1189/jlb.0313168. Epub 2013 Sep 9. J Leukoc Biol. 2014. PMID: 24018352
-
The ligation between ERMAP, galectin-9 and dectin-2 promotes Kupffer cell phagocytosis and antitumor immunity.Nat Immunol. 2023 Nov;24(11):1813-1824. doi: 10.1038/s41590-023-01634-7. Epub 2023 Oct 9. Nat Immunol. 2023. PMID: 37813965
-
Characterization of C-type lectins reveals an unexpectedly limited interaction between Cryptococcus neoformans spores and Dectin-1.PLoS One. 2017 Mar 10;12(3):e0173866. doi: 10.1371/journal.pone.0173866. eCollection 2017. PLoS One. 2017. PMID: 28282442 Free PMC article.
-
Targeting C-type lectin receptors with multivalent carbohydrate ligands.Adv Drug Deliv Rev. 2013 Aug;65(9):1271-81. doi: 10.1016/j.addr.2013.05.007. Epub 2013 May 30. Adv Drug Deliv Rev. 2013. PMID: 23727341 Review.
-
Systems biology of host-mycobiota interactions: dissecting Dectin-1 and Dectin-2 signalling in immune cells with DC-ATLAS.Immunobiology. 2013 Nov;218(11):1428-37. doi: 10.1016/j.imbio.2013.07.002. Epub 2013 Jul 13. Immunobiology. 2013. PMID: 23932568 Review.
Cited by
-
The nuclear factor ID3 endows macrophages with a potent anti-tumour activity.Nature. 2024 Feb;626(8000):864-873. doi: 10.1038/s41586-023-06950-4. Epub 2024 Feb 7. Nature. 2024. PMID: 38326607 Free PMC article.
-
Targeting macrophages in cancer immunotherapy.Signal Transduct Target Ther. 2021 Mar 26;6(1):127. doi: 10.1038/s41392-021-00506-6. Signal Transduct Target Ther. 2021. PMID: 33767177 Free PMC article.
-
Neem leaf glycoprotein binding to Dectin-1 receptors on dendritic cell induces type-1 immunity through CARD9 mediated intracellular signal to NFκB.Cell Commun Signal. 2024 Apr 23;22(1):237. doi: 10.1186/s12964-024-01576-z. Cell Commun Signal. 2024. PMID: 38649988 Free PMC article.
-
Tissue-Resident Macrophages in Cancer: Friend or Foe?Cancer Med. 2024 Nov;13(21):e70387. doi: 10.1002/cam4.70387. Cancer Med. 2024. PMID: 39494816 Free PMC article. Review.
-
Straight to the point: targeted mRNA-delivery to immune cells for improved vaccine design.Front Immunol. 2023 Nov 27;14:1294929. doi: 10.3389/fimmu.2023.1294929. eCollection 2023. Front Immunol. 2023. PMID: 38090568 Free PMC article. Review.
References
-
- Van den Eynden GG, et al. The multifaceted role of the microenvironment in liver metastasis: Biology and clinical implications. Cancer Res. 2013;73(7):2031–2043. - PubMed
-
- Protzer U, Maini MK, Knolle PA. Living in the liver: Hepatic infections. Nat Rev Immunol. 2012;12(3):201–213. - PubMed
-
- Heymann F, Tacke F. Immunology in the liver—from homeostasis to disease. Nat Rev Gastroenterol Hepatol. 2016;13(2):88–110. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

