PAR-CLIP and streamlined small RNA cDNA library preparation protocol for the identification of RNA binding protein target sites
- PMID: 27871973
- PMCID: PMC11145949
- DOI: 10.1016/j.ymeth.2016.11.009
PAR-CLIP and streamlined small RNA cDNA library preparation protocol for the identification of RNA binding protein target sites
Abstract
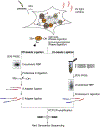
The study of protein-RNA interactions is critical for our understanding of cellular processes and regulatory circuits controlled by RNA binding proteins (RBPs). Recent next generation sequencing-based approaches significantly promoted our understanding of RNA biology and its importance for cell function. We present a streamlined protocol for Photoactivatable-Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation (PAR-CLIP), a technique that allows for the characterization of RBP binding sites on target RNAs at nucleotide resolution and transcriptome-wide scale. PAR-CLIP involves irreversible UV-mediated crosslinking of RNAs labeled with photoreactive nucleosides to interacting proteins, followed by stringent purification steps and the conversion of crosslinked RNA into small RNA cDNA libraries compatible with next-generation sequencing. The defining hallmark of PAR-CLIP is a diagnostic mutation at the crosslinking site that is introduced into cDNA during the library preparation process. This feature allows for efficient computational removal of contaminating sequences derived from non-crosslinked fragments of abundant cellular RNAs. In the following, we present two different step-by-step procedures for PAR-CLIP, which differ in the small RNA cDNA library preparation procedure: (1) Standard library preparation involving gel size selections after each enzymatic manipulation, and (2) A modified PAR-CLIP procedure ("on-beads" PAR-CLIP), where most RNA manipulations including the necessary adapter ligation steps are performed on the immobilized RNP. This streamlined procedure reduces the protocol preparation time by three days compared to the standard workflow.
Keywords: Next-generation sequencing; PAR-CLIP; Posttranscriptional Gene Regulation; RNA binding protein; Small RNA cDNA library.
Copyright © 2016. Published by Elsevier Inc.
Figures
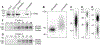

Similar articles
-
Optimization of PAR-CLIP for transcriptome-wide identification of binding sites of RNA-binding proteins.Methods. 2017 Apr 15;118-119:24-40. doi: 10.1016/j.ymeth.2016.10.007. Epub 2016 Oct 17. Methods. 2017. PMID: 27765618 Free PMC article. Review.
-
A non-radioactive, improved PAR-CLIP and small RNA cDNA library preparation protocol.Nucleic Acids Res. 2021 May 7;49(8):e45. doi: 10.1093/nar/gkab011. Nucleic Acids Res. 2021. PMID: 33503264 Free PMC article.
-
PAR-CliP--a method to identify transcriptome-wide the binding sites of RNA binding proteins.J Vis Exp. 2010 Jul 2;(41):2034. doi: 10.3791/2034. J Vis Exp. 2010. PMID: 20644507 Free PMC article.
-
Transcriptome-wide Identification of RNA-binding Protein Binding Sites Using Photoactivatable-Ribonucleoside-Enhanced Crosslinking Immunoprecipitation (PAR-CLIP).Curr Protoc Mol Biol. 2017 Apr 3;118:27.6.1-27.6.19. doi: 10.1002/cpmb.35. Curr Protoc Mol Biol. 2017. PMID: 28369676
-
Computational analysis of CLIP-seq data.Methods. 2017 Apr 15;118-119:60-72. doi: 10.1016/j.ymeth.2017.02.006. Epub 2017 Feb 22. Methods. 2017. PMID: 28254606 Review.
Cited by
-
Matrin3 regulates mitotic spindle dynamics by controlling alternative splicing of CDC14B.Cell Rep. 2023 Mar 28;42(3):112260. doi: 10.1016/j.celrep.2023.112260. Epub 2023 Mar 15. Cell Rep. 2023. PMID: 36924503 Free PMC article.
-
Probing Long Non-coding RNA-Protein Interactions.Front Mol Biosci. 2017 Jul 11;4:45. doi: 10.3389/fmolb.2017.00045. eCollection 2017. Front Mol Biosci. 2017. PMID: 28744458 Free PMC article. Review.
-
Epigenetic repression of antiviral genes by SARS-CoV-2 NSP1.PLoS One. 2024 Jan 26;19(1):e0297262. doi: 10.1371/journal.pone.0297262. eCollection 2024. PLoS One. 2024. PMID: 38277395 Free PMC article.
-
Impaired neurogenesis alters brain biomechanics in a neuroprogenitor-based genetic subtype of congenital hydrocephalus.Nat Neurosci. 2022 Apr;25(4):458-473. doi: 10.1038/s41593-022-01043-3. Epub 2022 Apr 4. Nat Neurosci. 2022. PMID: 35379995 Free PMC article.
-
Decoding the 5' nucleotide bias of PIWI-interacting RNAs.Nat Commun. 2019 Feb 19;10(1):828. doi: 10.1038/s41467-019-08803-z. Nat Commun. 2019. PMID: 30783109 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

