Histones Are Rapidly Loaded onto Unintegrated Retroviral DNAs Soon after Nuclear Entry
- PMID: 27866901
- PMCID: PMC5159289
- DOI: 10.1016/j.chom.2016.10.009
Histones Are Rapidly Loaded onto Unintegrated Retroviral DNAs Soon after Nuclear Entry
Abstract
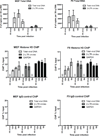
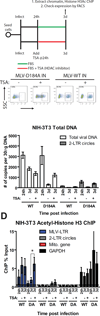
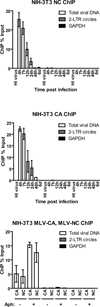
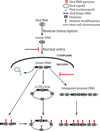
Chromosomal structure of nuclear DNA is usually maintained by insertion of nucleosomes into preexisting chromatin, both on newly synthesized DNA at replication forks and at sites of DNA damage. But during retrovirus infection, a histone-free DNA copy of the viral genome is synthesized that must be loaded with nucleosomes de novo. Here, we show that core histones are rapidly loaded onto unintegrated Moloney murine leukemia virus DNAs. Loading of nucleosomes requires nuclear entry, but does not require viral DNA integration. The histones associated with unintegrated DNAs become marked by covalent modifications, with a delay relative to the time of core histone loading. Expression from unintegrated DNA can be enhanced by modulation of the histone-modifying machinery. The data show that histone loading onto unintegrated DNAs occurs very rapidly after nuclear entry and does not require prior establishment of an integrated provirus.
Keywords: chromatin immunoprecipitation; epigenetics; histone; histone modifications; nucleosome; retrovirus.
Copyright © 2016 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Comment in
-
Viral infection: How histones go viral.Nat Rev Microbiol. 2016 Dec 9;15(1):3. doi: 10.1038/nrmicro.2016.186. Nat Rev Microbiol. 2016. PMID: 27932798 No abstract available.
Similar articles
-
Moloney Murine Leukemia Virus p12 Is Required for Histone Loading onto Retroviral DNAs.J Virol. 2021 Jul 12;95(15):e0049521. doi: 10.1128/JVI.00495-21. Epub 2021 Jul 12. J Virol. 2021. PMID: 34011543 Free PMC article.
-
Unintegrated HIV-1 DNAs are loaded with core and linker histones and transcriptionally silenced.Proc Natl Acad Sci U S A. 2019 Nov 19;116(47):23735-23742. doi: 10.1073/pnas.1912638116. Epub 2019 Nov 4. Proc Natl Acad Sci U S A. 2019. PMID: 31685613 Free PMC article.
-
Silencing of Unintegrated Retroviral DNAs.Viruses. 2021 Nov 9;13(11):2248. doi: 10.3390/v13112248. Viruses. 2021. PMID: 34835055 Free PMC article. Review.
-
CHAF1A/B mediate silencing of unintegrated HIV-1 DNAs early in infection.Proc Natl Acad Sci U S A. 2022 Jan 25;119(4):e2116735119. doi: 10.1073/pnas.2116735119. Proc Natl Acad Sci U S A. 2022. PMID: 35074917 Free PMC article.
-
H3-H4 Histone Chaperone Pathways.Annu Rev Genet. 2018 Nov 23;52:109-130. doi: 10.1146/annurev-genet-120417-031547. Epub 2018 Sep 5. Annu Rev Genet. 2018. PMID: 30183406 Review.
Cited by
-
Viruses in the Nucleus.Cold Spring Harb Perspect Biol. 2021 Aug 2;13(8):a039446. doi: 10.1101/cshperspect.a039446. Cold Spring Harb Perspect Biol. 2021. PMID: 33753405 Free PMC article. Review.
-
How cells hush a viral invader.Nature. 2018 Dec;564(7735):193-194. doi: 10.1038/d41586-018-07493-9. Nature. 2018. PMID: 30531877 No abstract available.
-
POLE3 is a repressor of unintegrated HIV-1 DNA required for efficient virus integration and escape from innate immune sensing.Sci Adv. 2023 Nov 3;9(44):eadh3642. doi: 10.1126/sciadv.adh3642. Epub 2023 Nov 3. Sci Adv. 2023. PMID: 37922361 Free PMC article.
-
Prasinovirus Attack of Ostreococcus Is Furtive by Day but Savage by Night.J Virol. 2018 Jan 30;92(4):e01703-17. doi: 10.1128/JVI.01703-17. Print 2018 Feb 15. J Virol. 2018. PMID: 29187539 Free PMC article.
-
CRL4-DCAF1 Ubiquitin Ligase Dependent Functions of HIV Viral Protein R and Viral Protein X.Viruses. 2024 Aug 17;16(8):1313. doi: 10.3390/v16081313. Viruses. 2024. PMID: 39205287 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

