CRISPR/Cas9-Induced Double-Strand Break Repair in Arabidopsis Nonhomologous End-Joining Mutants
- PMID: 27866150
- PMCID: PMC5217109
- DOI: 10.1534/g3.116.035204
CRISPR/Cas9-Induced Double-Strand Break Repair in Arabidopsis Nonhomologous End-Joining Mutants
Abstract
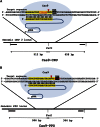
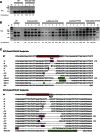
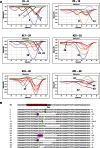
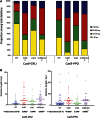
Double-strand breaks (DSBs) are one of the most harmful DNA lesions. Cells utilize two main pathways for DSB repair: homologous recombination (HR) and nonhomologous end-joining (NHEJ). NHEJ can be subdivided into the KU-dependent classical NHEJ (c-NHEJ) and the more error-prone KU-independent backup-NHEJ (b-NHEJ) pathways, involving the poly (ADP-ribose) polymerases (PARPs). However, in the absence of these factors, cells still seem able to adequately maintain genome integrity, suggesting the presence of other b-NHEJ repair factors or pathways independent from KU and PARPs. The outcome of DSB repair by NHEJ pathways can be investigated by using artificial sequence-specific nucleases such as CRISPR/Cas9 to induce DSBs at a target of interest. Here, we used CRISPR/Cas9 for DSB induction at the Arabidopsis cruciferin 3 (CRU3) and protoporphyrinogen oxidase (PPO) genes. DSB repair outcomes via NHEJ were analyzed using footprint analysis in wild-type plants and plants deficient in key factors of c-NHEJ (ku80), b-NHEJ (parp1 parp2), or both (ku80 parp1 parp2). We found that larger deletions of >20 bp predominated after DSB repair in ku80 and ku80 parp1 parp2 mutants, corroborating with a role of KU in preventing DSB end resection. Deletion lengths did not significantly differ between ku80 and ku80 parp1 parp2 mutants, suggesting that a KU- and PARP-independent b-NHEJ mechanism becomes active in these mutants. Furthermore, microhomologies and templated insertions were observed at the repair junctions in the wild type and all mutants. Since these characteristics are hallmarks of polymerase θ-mediated DSB repair, we suggest a possible role for this recently discovered polymerase in DSB repair in plants.
Keywords: Arabidopsis thaliana; CRISPR/Cas9; KU80; double-strand break; nonhomologous end-joining.
Copyright © 2017 Shen et al.
Figures

Similar articles
-
Common and unique genetic interactions of the poly(ADP-ribose) polymerases PARP1 and PARP2 with DNA double-strand break repair pathways.DNA Repair (Amst). 2016 Sep;45:56-62. doi: 10.1016/j.dnarep.2016.06.001. Epub 2016 Jun 16. DNA Repair (Amst). 2016. PMID: 27373144 Free PMC article.
-
Analysis of chromatid-break-repair detects a homologous recombination to non-homologous end-joining switch with increasing load of DNA double-strand breaks.Mutat Res Genet Toxicol Environ Mutagen. 2021 Jul;867:503372. doi: 10.1016/j.mrgentox.2021.503372. Epub 2021 Jun 12. Mutat Res Genet Toxicol Environ Mutagen. 2021. PMID: 34266628
-
Poly(ADP-ribose)polymerases are involved in microhomology mediated back-up non-homologous end joining in Arabidopsis thaliana.Plant Mol Biol. 2013 Jul;82(4-5):339-51. doi: 10.1007/s11103-013-0065-9. Epub 2013 Apr 28. Plant Mol Biol. 2013. PMID: 23625359
-
DNA Repair Pathway Choices in CRISPR-Cas9-Mediated Genome Editing.Trends Genet. 2021 Jul;37(7):639-656. doi: 10.1016/j.tig.2021.02.008. Epub 2021 Apr 22. Trends Genet. 2021. PMID: 33896583 Free PMC article. Review.
-
Mechanisms of DNA double strand break repair and chromosome aberration formation.Cytogenet Genome Res. 2004;104(1-4):14-20. doi: 10.1159/000077461. Cytogenet Genome Res. 2004. PMID: 15162010 Review.
Cited by
-
The SAP domain of Ku facilitates its efficient loading onto DNA ends.Nucleic Acids Res. 2023 Nov 27;51(21):11706-11716. doi: 10.1093/nar/gkad850. Nucleic Acids Res. 2023. PMID: 37850645 Free PMC article.
-
CRISPR/Cas9 disruption of UGT71L1 in poplar connects salicinoid and salicylic acid metabolism and alters growth and morphology.Plant Cell. 2022 Jul 30;34(8):2925-2947. doi: 10.1093/plcell/koac135. Plant Cell. 2022. PMID: 35532172 Free PMC article.
-
CRISPR-based tools for plant genome engineering.Emerg Top Life Sci. 2017 Nov 10;1(2):135-149. doi: 10.1042/ETLS20170011. Emerg Top Life Sci. 2017. PMID: 33525768 Free PMC article.
-
Detection and characterization of genome-wide mutations in M1 vegetative cells of gamma-irradiated Arabidopsis.PLoS Genet. 2022 Jan 20;18(1):e1009979. doi: 10.1371/journal.pgen.1009979. eCollection 2022 Jan. PLoS Genet. 2022. PMID: 35051177 Free PMC article.
-
Poly(ADP-Ribose) Polymerases in Plants and Their Human Counterparts: Parallels and Peculiarities.Int J Mol Sci. 2019 Apr 2;20(7):1638. doi: 10.3390/ijms20071638. Int J Mol Sci. 2019. PMID: 30986964 Free PMC article. Review.
References
-
- Alonso J. M., Stepanova A. N., Leisse T. J., Kim C. J., Chen H., et al. , 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. - PubMed
-
- Belhaj K., Chaparro-Garcia A., Kamoun S., Patron N. J., Nekrasov V., 2015. Editing plant genomes with CRISPR/Cas9. Curr. Opin. Biotechnol. 32: 76–84. - PubMed
-
- Clough S. J., Bent A. F., 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. - PubMed
-
- Deriano L., Roth D. B., 2013. Modernizing the nonhomologous end-joining repertoire: alternative and classical NHEJ share the stage. Annu. Rev. Genet. 47: 433–455. - PubMed
-
- de Pater S., Neuteboom L. W., Pinas J. E., Hooykaas P. J. J., van der Zaal B. J., 2009. ZFN-induced mutagenesis and gene-targeting in Arabidopsis through Agrobacterium-mediated floral dip transformation. Plant Biotechnol. J. 7: 821–835. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
