Splicing repression allows the gradual emergence of new Alu-exons in primate evolution
- PMID: 27861119
- PMCID: PMC5115870
- DOI: 10.7554/eLife.19545
Splicing repression allows the gradual emergence of new Alu-exons in primate evolution
Abstract
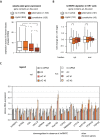
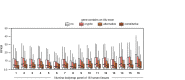
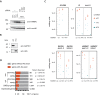
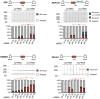
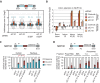
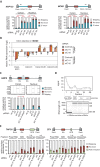
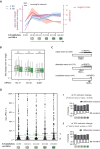
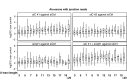
Alu elements are retrotransposons that frequently form new exons during primate evolution. Here, we assess the interplay of splicing repression by hnRNPC and nonsense-mediated mRNA decay (NMD) in the quality control and evolution of new Alu-exons. We identify 3100 new Alu-exons and show that NMD more efficiently recognises transcripts with Alu-exons compared to other exons with premature termination codons. However, some Alu-exons escape NMD, especially when an adjacent intron is retained, highlighting the importance of concerted repression by splicing and NMD. We show that evolutionary progression of 3' splice sites is coupled with longer repressive uridine tracts. Once the 3' splice site at ancient Alu-exons reaches a stable phase, splicing repression by hnRNPC decreases, but the exons generally remain sensitive to NMD. We conclude that repressive motifs are strongest next to cryptic exons and that gradual weakening of these motifs contributes to the evolutionary emergence of new alternative exons.
Keywords: Alu elements; evolutionary biology; exon evolution; genomics; human; nonsense mediated mRNA decay; pre-mRNA processing; splicing fidelity.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures



Similar articles
-
Integrative transcriptomic analysis suggests new autoregulatory splicing events coupled with nonsense-mediated mRNA decay.Nucleic Acids Res. 2019 Jun 4;47(10):5293-5306. doi: 10.1093/nar/gkz193. Nucleic Acids Res. 2019. PMID: 30916337 Free PMC article.
-
Intergenic Alu exonisation facilitates the evolution of tissue-specific transcript ends.Nucleic Acids Res. 2015 Dec 2;43(21):10492-505. doi: 10.1093/nar/gkv956. Epub 2015 Sep 22. Nucleic Acids Res. 2015. PMID: 26400176 Free PMC article.
-
Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements.Cell. 2013 Jan 31;152(3):453-66. doi: 10.1016/j.cell.2012.12.023. Cell. 2013. PMID: 23374342 Free PMC article.
-
Exonization of transposed elements: A challenge and opportunity for evolution.Biochimie. 2011 Nov;93(11):1928-34. doi: 10.1016/j.biochi.2011.07.014. Epub 2011 Jul 26. Biochimie. 2011. PMID: 21787833 Review.
-
The role of alternative splicing coupled to nonsense-mediated mRNA decay in human disease.Int J Biochem Cell Biol. 2017 Oct;91(Pt B):168-175. doi: 10.1016/j.biocel.2017.07.013. Epub 2017 Jul 22. Int J Biochem Cell Biol. 2017. PMID: 28743674 Review.
Cited by
-
Critical Differential Expression Assessment for Individual Bulk RNA-Seq Projects.bioRxiv [Preprint]. 2024 Feb 12:2024.02.10.579728. doi: 10.1101/2024.02.10.579728. bioRxiv. 2024. PMID: 38405814 Free PMC article. Preprint.
-
Alternative splicing as a source of phenotypic diversity.Nat Rev Genet. 2022 Nov;23(11):697-710. doi: 10.1038/s41576-022-00514-4. Epub 2022 Jul 12. Nat Rev Genet. 2022. PMID: 35821097 Review.
-
Human Survival Motor Neuron genes generate a vast repertoire of circular RNAs.Nucleic Acids Res. 2019 Apr 8;47(6):2884-2905. doi: 10.1093/nar/gkz034. Nucleic Acids Res. 2019. PMID: 30698797 Free PMC article.
-
Multilayered control of exon acquisition permits the emergence of novel forms of regulatory control.Genome Biol. 2019 Jul 17;20(1):141. doi: 10.1186/s13059-019-1757-5. Genome Biol. 2019. PMID: 31315652 Free PMC article.
-
A PPARG Splice Variant in Granulosa Cells Is Associated with Polycystic Ovary Syndrome.J Clin Med. 2022 Dec 8;11(24):7285. doi: 10.3390/jcm11247285. J Clin Med. 2022. PMID: 36555903 Free PMC article.
References
-
- Amit M, Donyo M, Hollander D, Goren A, Kim E, Gelfman S, Lev-Maor G, Burstein D, Schwartz S, Postolsky B, Pupko T, Ast G. Differential GC content between exons and introns establishes distinct strategies of splice-site recognition. Cell Reports. 2012;1:543–556. doi: 10.1016/j.celrep.2012.03.013. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

