Activation of transcription enforces the formation of distinct nuclear bodies in zebrafish embryos
- PMID: 27858508
- PMCID: PMC5519242
- DOI: 10.1080/15476286.2016.1255397
Activation of transcription enforces the formation of distinct nuclear bodies in zebrafish embryos
Abstract
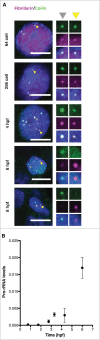
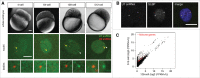
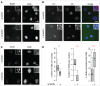
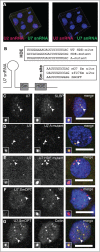
Nuclear bodies are cellular compartments that lack lipid bilayers and harbor specific RNAs and proteins. Recent proposals that nuclear bodies form through liquid-liquid phase separation leave the question of how different nuclear bodies maintain their distinct identities unanswered. Here we investigate Cajal bodies (CBs), histone locus bodies (HLBs) and nucleoli - involved in assembly of the splicing machinery, histone mRNA 3' end processing, and rRNA processing, respectively - in the embryos of the zebrafish, Danio rerio. We take advantage of the transcriptional silence of the 1-cell embryo and follow nuclear body appearance as zygotic transcription becomes activated. CBs are present from fertilization onwards, while HLB and nucleolar components formed foci several hours later when histone genes and rDNA became active. HLB formation was blocked by transcription inhibition, suggesting nascent histone transcripts recruit HLB components like U7 snRNP. Surprisingly, we found that U7 base-pairing with nascent histone transcripts was not required for localization to HLBs. Rather, the type of Sm ring assembled on U7 determined its targeting to HLBs or CBs; the spliceosomal Sm ring targeted snRNAs to CBs while the specialized U7 Sm-ring localized to HLBs, demonstrating the contribution of protein constituents to the distinction among nuclear bodies. Thus, nucleolar, HLB, and CB components can mix in early embryogenesis when transcription is naturally or artificially silenced. These data support a model in which transcription of specific gene loci nucleates nuclear body components with high specificity and fidelity to perform distinct regulatory functions.
Keywords: Cajal body; coilin; fibrillarin; histone locus body; liquid phase separation; zebrafish; zygotic genome activation.
Figures
Similar articles
-
Dynamics and Function of Nuclear Bodies during Embryogenesis.Biochemistry. 2018 May 1;57(17):2462-2469. doi: 10.1021/acs.biochem.7b01262. Epub 2018 Mar 7. Biochemistry. 2018. PMID: 29473743 Review.
-
Dynamic control of Cajal body number during zebrafish embryogenesis.Nucleus. 2010 Jan-Feb;1(1):96-108. doi: 10.4161/nucl.1.1.10680. Nucleus. 2010. PMID: 21327108 Free PMC article.
-
Specific genomic cues regulate Cajal body assembly.RNA Biol. 2017 Jun 3;14(6):791-803. doi: 10.1080/15476286.2016.1243648. Epub 2016 Oct 7. RNA Biol. 2017. PMID: 27715441 Free PMC article. Review.
-
The Drosophila melanogaster Cajal body.J Cell Biol. 2006 Mar 13;172(6):875-84. doi: 10.1083/jcb.200511038. J Cell Biol. 2006. PMID: 16533947 Free PMC article.
-
Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product.J Cell Biol. 2001 Jul 23;154(2):293-307. doi: 10.1083/jcb.200104083. J Cell Biol. 2001. PMID: 11470819 Free PMC article.
Cited by
-
The role of transcription in shaping the spatial organization of the genome.Nat Rev Mol Cell Biol. 2019 Jun;20(6):327-337. doi: 10.1038/s41580-019-0114-6. Nat Rev Mol Cell Biol. 2019. PMID: 30886333 Free PMC article. Review.
-
Physiology and pharmacological targeting of phase separation.J Biomed Sci. 2024 Jan 20;31(1):11. doi: 10.1186/s12929-024-00993-z. J Biomed Sci. 2024. PMID: 38245749 Free PMC article. Review.
-
SGF29 nuclear condensates reinforce cellular aging.Cell Discov. 2023 Nov 7;9(1):110. doi: 10.1038/s41421-023-00602-7. Cell Discov. 2023. PMID: 37935676 Free PMC article.
-
A dual ribosomal system in the zebrafish soma and germline.bioRxiv [Preprint]. 2024 Aug 30:2024.08.29.610041. doi: 10.1101/2024.08.29.610041. bioRxiv. 2024. PMID: 39257781 Free PMC article. Preprint.
-
The dynamics of three-dimensional chromatin organization and phase separation in cell fate transitions and diseases.Cell Regen. 2022 Dec 21;11(1):42. doi: 10.1186/s13619-022-00145-4. Cell Regen. 2022. PMID: 36539553 Free PMC article. Review.
References
-
- Mao YS, Zhang B, Spector DL. Biogenesis and function of nuclear bodies. Trends in genetics : TIG 2011; 27(8):p. 295-306; PMID:21680045; https://doi.org/10.1016/j.tig.2011.05.006 - DOI - PMC - PubMed
-
- Courchaine EM, Lu A, Neugebauer KM. Droplet organelles? EMBO J 2016; 35(15):p. 1603-12; PMID:27357569; https://doi.org/10.15252/embj.201593517 - DOI - PMC - PubMed
-
- Matera AG, Izaguire-Sierra M, Praveen K, Rajendra TK. Nuclear bodies: random aggregates of sticky proteins or crucibles of macromolecular assembly? Developmental cell 2009; 17(5):p. 639-47; PMID:19922869; https://doi.org/10.1016/j.devcel.2009.10.017 - DOI - PMC - PubMed
-
- Berry J, Weber SC, Vaidya N, Haataja M, Brangwynne CP. RNA transcription modulates phase transition-driven nuclear body assembly. Proc Natl Acad Sci U S A 2015; 112(38):p. E5237-45; PMID:26351690; https://doi.org/10.1073/pnas.1509317112 - DOI - PMC - PubMed
-
- Falahati H, Pelham-Webb B, Blythe S, Wieschaus E. Nucleation by rRNA Dictates the Precision of Nucleolus Assembly. Curr Biol 2016; 26(3):p. 277-85; PMID:26776729; https://doi.org/10.1016/j.cub.2015.11.065 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
