A core phylogeny of Dictyostelia inferred from genomes representative of the eight major and minor taxonomic divisions of the group
- PMID: 27855631
- PMCID: PMC5114724
- DOI: 10.1186/s12862-016-0825-7
A core phylogeny of Dictyostelia inferred from genomes representative of the eight major and minor taxonomic divisions of the group
Abstract
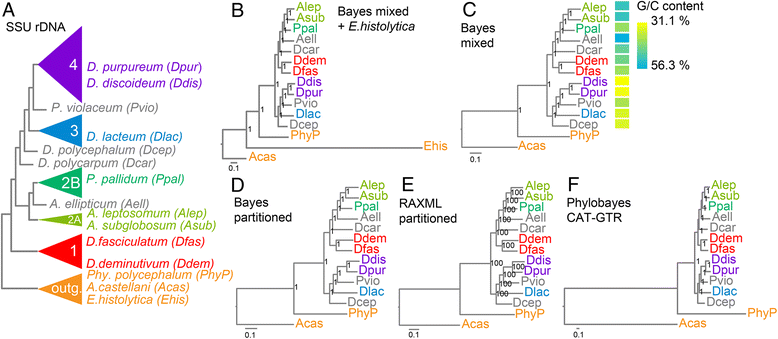
Background: Dictyostelia are a well-studied group of organisms with colonial multicellularity, which are members of the mostly unicellular Amoebozoa. A phylogeny based on SSU rDNA data subdivided all Dictyostelia into four major groups, but left the position of the root and of six group-intermediate taxa unresolved. Recent phylogenies inferred from 30 or 213 proteins from sequenced genomes, positioned the root between two branches, each containing two major groups, but lacked data to position the group-intermediate taxa. Since the positions of these early diverging taxa are crucial for understanding the evolution of phenotypic complexity in Dictyostelia, we sequenced six representative genomes of early diverging taxa.
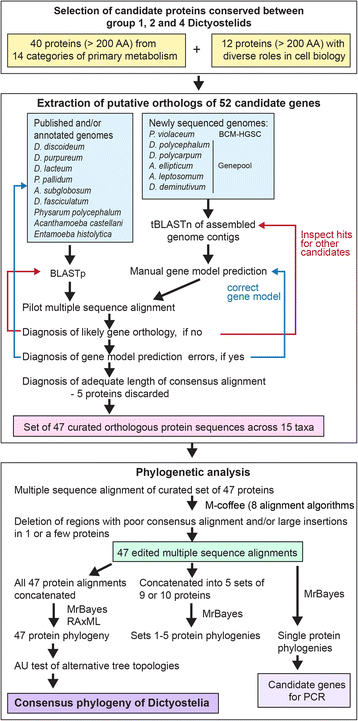
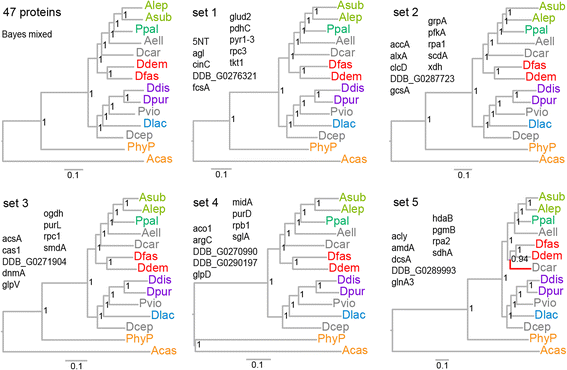
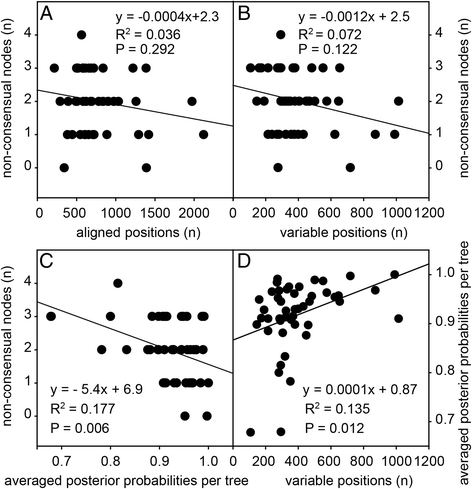
Results: We retrieved orthologs of 47 housekeeping proteins with an average size of 890 amino acids from six newly sequenced and eight published genomes of Dictyostelia and unicellular Amoebozoa and inferred phylogenies from single and concatenated protein sequence alignments. Concatenated alignments of all 47 proteins, and four out of five subsets of nine concatenated proteins all produced the same consensus phylogeny with 100% statistical support. Trees inferred from just two out of the 47 proteins, individually reproduced the consensus phylogeny, highlighting that single gene phylogenies will rarely reflect correct species relationships. However, sets of two or three concatenated proteins again reproduced the consensus phylogeny, indicating that a small selection of genes suffices for low cost classification of as yet unincorporated or newly discovered dictyostelid and amoebozoan taxa by gene amplification.
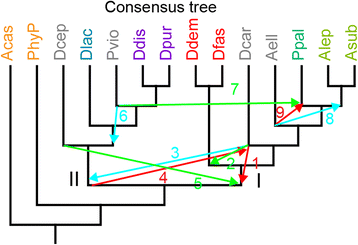
Conclusions: The multi-locus consensus phylogeny shows that groups 1 and 2 are sister clades in branch I, with the group-intermediate taxon D. polycarpum positioned as outgroup to group 2. Branch II consists of groups 3 and 4, with the group-intermediate taxon Polysphondylium violaceum positioned as sister to group 4, and the group-intermediate taxon Dictyostelium polycephalum branching at the base of that whole clade. Given the data, the approximately unbiased test rejects all alternative topologies favoured by SSU rDNA and individual proteins with high statistical support. The test also rejects monophyletic origins for the genera Acytostelium, Polysphondylium and Dictyostelium. The current position of Acytostelium ellipticum in the consensus phylogeny indicates that somatic cells were lost twice in Dictyostelia.
Keywords: Dictyostelia; Evolution of multicellularity; Evolution of soma; Multi-locus phylogeny; Phylogenomics; Taxonomy.
Figures
Similar articles
-
A well supported multi gene phylogeny of 52 dictyostelia.Mol Phylogenet Evol. 2019 May;134:66-73. doi: 10.1016/j.ympev.2019.01.017. Epub 2019 Jan 31. Mol Phylogenet Evol. 2019. PMID: 30711536 Free PMC article.
-
Root of Dictyostelia based on 213 universal proteins.Mol Phylogenet Evol. 2015 Nov;92:53-62. doi: 10.1016/j.ympev.2015.05.017. Epub 2015 Jun 3. Mol Phylogenet Evol. 2015. PMID: 26048704
-
A Deep Hidden Diversity of Dictyostelia.Protist. 2018 Feb;169(1):64-78. doi: 10.1016/j.protis.2017.12.005. Epub 2018 Jan 9. Protist. 2018. PMID: 29427837
-
Evolution and diversity of dictyostelid social amoebae.Protist. 2012 May;163(3):327-43. doi: 10.1016/j.protis.2011.09.004. Epub 2011 Dec 30. Protist. 2012. PMID: 22209334 Review.
-
Evolution of multicellularity in Dictyostelia.Int J Dev Biol. 2019;63(8-9-10):359-369. doi: 10.1387/ijdb.190108ps. Int J Dev Biol. 2019. PMID: 31840775 Free PMC article. Review.
Cited by
-
Evolution of Multicellular Complexity in The Dictyostelid Social Amoebas.Genes (Basel). 2021 Mar 27;12(4):487. doi: 10.3390/genes12040487. Genes (Basel). 2021. PMID: 33801615 Free PMC article. Review.
-
Diversity of Dictyostelid Cellular Slime Molds, Including Two Species New to Science, in Forest Soils of Changbai Mountain, China.Microbiol Spectr. 2022 Oct 26;10(5):e0240222. doi: 10.1128/spectrum.02402-22. Epub 2022 Oct 3. Microbiol Spectr. 2022. PMID: 36190423 Free PMC article.
-
A well supported multi gene phylogeny of 52 dictyostelia.Mol Phylogenet Evol. 2019 May;134:66-73. doi: 10.1016/j.ympev.2019.01.017. Epub 2019 Jan 31. Mol Phylogenet Evol. 2019. PMID: 30711536 Free PMC article.
-
Chromosome-level genome assembly and annotation of the social amoeba Dictyostelium firmibasis.Sci Data. 2024 Jun 22;11(1):678. doi: 10.1038/s41597-024-03513-8. Sci Data. 2024. PMID: 38909042 Free PMC article.
-
Phylogeny-wide conservation and change in developmental expression, cell-type specificity and functional domains of the transcriptional regulators of social amoebas.BMC Genomics. 2019 Nov 21;20(1):890. doi: 10.1186/s12864-019-6239-3. BMC Genomics. 2019. PMID: 31752673 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

