IGFBP2 expression predicts IDH-mutant glioma patient survival
- PMID: 27852048
- PMCID: PMC5352106
- DOI: 10.18632/oncotarget.13329
IGFBP2 expression predicts IDH-mutant glioma patient survival
Abstract
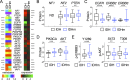
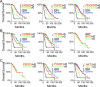
Mutations of the isocitrate dehydrogenase (IDH) 1 and 2 genes occur in ~80% of lower-grade (WHO grade II and grade III) gliomas. Mutant IDH produces (R)-2-hydroxyglutarate, which induces DNA hypermethylation and presumably drives tumorigenesis. Interestingly, IDH mutations are associated with improved survival in glioma patients, but the underlying mechanism for the difference in survival remains unclear. Through comparative analyses of 286 cases of IDH-wildtype and IDH-mutant lower-grade glioma from a TCGA data set, we report that IDH-mutant gliomas have increased expression of tumor-suppressor genes (NF1, PTEN, and PIK3R1) and decreased expression of oncogenes(AKT2, ARAF, ERBB2, FGFR3, and PDGFRB) and glioma progression genes (FOXM1, IGFBP2, and WWTR1) compared with IDH-wildtype gliomas. Furthermore, each of these genes is prognostic in overall gliomas; however, within the IDH-mutant group, none remains prognostic except IGFBP2 (encodinginsulin-like growth factor binding protein 2). Through validation in an independent cohort, we show that patients with low IGFBP2 expressiondisplay a clear advantage in overall and disease-free survival, whereas those with high IGFBP2 expressionhave worse median survival than IDH-wildtype patients. These observations hold true across different histological and molecular subtypes of lower-grade glioma. We propose therefore that an unexpected biological consequence of IDH mutations in glioma is to ameliorate patient survival by promoting tumor-suppressor signaling while inhibiting that of oncogenes, particularly IGFBP2.
Keywords: DNA hypermethylation; IDH; IGFBP2; glioma; prognosis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
SHOX2 is a Potent Independent Biomarker to Predict Survival of WHO Grade II-III Diffuse Gliomas.EBioMedicine. 2016 Nov;13:80-89. doi: 10.1016/j.ebiom.2016.10.040. Epub 2016 Oct 28. EBioMedicine. 2016. PMID: 27840009 Free PMC article.
-
Prognostic role of mitochondrial pyruvate carrier in isocitrate dehydrogenase-mutant glioma.J Neurosurg. 2019 Jan 1;130(1):56-66. doi: 10.3171/2017.9.JNS172036. J Neurosurg. 2019. PMID: 29547090
-
TERT promoter mutation and its interaction with IDH mutations in glioma: Combined TERT promoter and IDH mutations stratifies lower-grade glioma into distinct survival subgroups-A meta-analysis of aggregate data.Crit Rev Oncol Hematol. 2017 Dec;120:1-9. doi: 10.1016/j.critrevonc.2017.09.013. Epub 2017 Oct 3. Crit Rev Oncol Hematol. 2017. PMID: 29198322 Review.
-
PI3 kinase mutations and mutational load as poor prognostic markers in diffuse glioma patients.Acta Neuropathol Commun. 2015 Dec 23;3:88. doi: 10.1186/s40478-015-0265-4. Acta Neuropathol Commun. 2015. PMID: 26699864 Free PMC article.
-
Isocitrate dehydrogenase-mutant glioma: Evolving clinical and therapeutic implications.Cancer. 2017 Dec 1;123(23):4535-4546. doi: 10.1002/cncr.31039. Epub 2017 Oct 5. Cancer. 2017. PMID: 28980701 Review.
Cited by
-
Multivariate analysis reveals differentially expressed genes among distinct subtypes of diffuse astrocytic gliomas: diagnostic implications.Sci Rep. 2020 Jul 9;10(1):11270. doi: 10.1038/s41598-020-67743-7. Sci Rep. 2020. PMID: 32647207 Free PMC article.
-
Multiparametric MR radiomics in brain glioma: models comparation to predict biomarker status.BMC Med Imaging. 2022 Aug 5;22(1):137. doi: 10.1186/s12880-022-00865-8. BMC Med Imaging. 2022. PMID: 35931979 Free PMC article.
-
Friend or foe-IDH1 mutations in glioma 10 years on.Carcinogenesis. 2019 Nov 25;40(11):1299-1307. doi: 10.1093/carcin/bgz134. Carcinogenesis. 2019. PMID: 31504231 Free PMC article. Review.
-
Expanding the scope of candidate prognostic marker IGFBP2 in glioblastoma.Biosci Rep. 2019 Jul 18;39(7):BSR20190770. doi: 10.1042/BSR20190770. Print 2019 Jul 31. Biosci Rep. 2019. PMID: 31296788 Free PMC article.
-
The neural stem-cell marker CD24 is specifically upregulated in IDH-mutant glioma.Transl Oncol. 2020 Oct;13(10):100819. doi: 10.1016/j.tranon.2020.100819. Epub 2020 Jul 1. Transl Oncol. 2020. PMID: 32622311 Free PMC article.
References
-
- Ostrom QT, Gittleman H, Liao P, Rouse C, Chen Y, Dowling J, Wolinsky Y, Kruchko C, Barnholtz-Sloan J. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2007-2011. Neuro-oncology. 2014;16(Suppl 4):iv1–63. doi: 10.1093/neuonc/nou223. - DOI - PMC - PubMed
-
- Bai H, Harmancı AS, Erson-Omay EZ, Li J, Coşkun S, Simon M, Krischek B, Özduman K, Omay SB, Sorensen EA, Turcan S, Bakırcığlu M, Carrión-Grant G, et al. Integrated genomic characterization of IDH1-mutant glioma malignant progression. Nat Genet. 2016;48:59–66. doi: 10.1038/ng.3457. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

