Analysis of Microarray Data on Gene Expression and Methylation to Identify Long Non-coding RNAs in Non-small Cell Lung Cancer
- PMID: 27849024
- PMCID: PMC5110979
- DOI: 10.1038/srep37233
Analysis of Microarray Data on Gene Expression and Methylation to Identify Long Non-coding RNAs in Non-small Cell Lung Cancer
Abstract
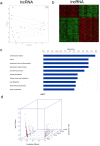
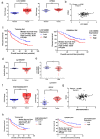
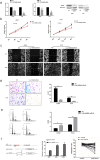
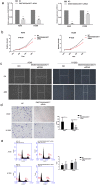
To identify what long non-coding RNAs (lncRNAs) are involved in non-small cell lung cancer (NSCLC), we analyzed microarray data on gene expression and methylation. Gene expression chip and HumanMethylation450BeadChip were used to interrogate genome-wide expression and methylation in tumor samples. Differential expression and methylation were analyzed through comparing tumors with adjacent non-tumor tissues. LncRNAs expressed differentially and correlated with coding genes and DNA methylation were validated in additional tumor samples using RT-qPCR and pyrosequencing. In vitro experiments were performed to evaluate lncRNA's effects on tumor cells. We identified 8,500 lncRNAs expressed differentially between tumor and non-tumor tissues, of which 1,504 were correlated with mRNA expression. Two of the lncRNAs, LOC146880 and ENST00000439577, were positively correlated with expression of two cancer-related genes, KPNA2 and RCC2, respectively. High expression of LOC146880 and ENST00000439577 were also associated with poor survival. Analysis of lncRNA expression in relation to DNA methylation showed that LOC146880 expression was down-regulated by DNA methylation in its promoter. Lowering the expression of LOC146880 or ENST00000439577 in tumor cells could inhibit cell proliferation, invasion and migration. Analysis of microarray data on gene expression and methylation allows us to identify two lncRNAs, LOC146880 and ENST00000439577, which may promote the progression of NSCLC.
Figures
Similar articles
-
RNA-Seq analysis of non-small cell lung cancer in female never-smokers reveals candidate cancer-associated long non-coding RNAs.Pathol Res Pract. 2016 Jun;212(6):549-54. doi: 10.1016/j.prp.2016.03.006. Epub 2016 Mar 19. Pathol Res Pract. 2016. PMID: 27067812
-
Study on expression of lncRNA RGMB-AS1 and repulsive guidance molecule b in non-small cell lung cancer.Diagn Pathol. 2015 Jun 9;10:63. doi: 10.1186/s13000-015-0297-x. Diagn Pathol. 2015. PMID: 26055877 Free PMC article.
-
Integrative CAGE and DNA Methylation Profiling Identify Epigenetically Regulated Genes in NSCLC.Mol Cancer Res. 2017 Oct;15(10):1354-1365. doi: 10.1158/1541-7786.MCR-17-0191. Epub 2017 Jul 11. Mol Cancer Res. 2017. PMID: 28698358
-
[Progress of Long Non-coding RNAs in Non-small Cell Lung Cancer].Zhongguo Fei Ai Za Zhi. 2016 Feb;19(2):108-12. doi: 10.3779/j.issn.1009-3419.2016.02.10. Zhongguo Fei Ai Za Zhi. 2016. PMID: 26903166 Free PMC article. Review. Chinese.
-
Long noncoding RNAs: new insights into non-small cell lung cancer biology, diagnosis and therapy.Med Oncol. 2016 Feb;33(2):18. doi: 10.1007/s12032-016-0731-2. Epub 2016 Jan 19. Med Oncol. 2016. PMID: 26786153 Review.
Cited by
-
LncRNA LOC146880 promotes esophageal squamous cell carcinoma progression via miR-328-5p/FSCN1/MAPK axis.Aging (Albany NY). 2021 May 18;13(10):14198-14218. doi: 10.18632/aging.203037. Epub 2021 May 18. Aging (Albany NY). 2021. PMID: 34016787 Free PMC article.
-
Methylation and transcriptome analysis reveal lung adenocarcinoma-specific diagnostic biomarkers.J Transl Med. 2019 Sep 27;17(1):324. doi: 10.1186/s12967-019-2068-z. J Transl Med. 2019. PMID: 31558162 Free PMC article.
-
Long non-coding RNA dysregulation is a frequent event in non-small cell lung carcinoma pathogenesis.Br J Cancer. 2020 Mar;122(7):1050-1058. doi: 10.1038/s41416-020-0742-9. Epub 2020 Feb 5. Br J Cancer. 2020. PMID: 32020063 Free PMC article.
-
Comprehensive analysis of prognostic value and immune infiltration of Regulator of Chromosome Condensation 2 in lung adenocarcinoma.J Cancer. 2024 Feb 11;15(7):1901-1915. doi: 10.7150/jca.91367. eCollection 2024. J Cancer. 2024. PMID: 38434981 Free PMC article.
-
New Perspectives in Different Gene Expression Profiles for Early and Locally Advanced Non-Small Cell Lung Cancer Stem Cells.Front Oncol. 2021 Mar 31;11:613198. doi: 10.3389/fonc.2021.613198. eCollection 2021. Front Oncol. 2021. PMID: 33868998 Free PMC article.
References
-
- Ferlay J. et al.. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 [Internet]. Lyon, France: International Agency for Research on Cancer. (2013) Available at: http://globocan.iarc.fr. (Accessed: 4th November 2015).
-
- Heller G., Zielinski C. C. & Zöchbauer-Müller S. Lung cancer: From single-gene methylation to methylome profiling. Cancer Metastasis Rev. 29, 95–107 (2010). - PubMed
-
- Walter K. et al.. DNA methylation profiling defines clinically relevant biological subsets of non-small cell lung cancer. Clin Cancer Res. 18, 2360–2373 (2012). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

