Overcoming a nucleosomal barrier to replication
- PMID: 27847876
- PMCID: PMC5106197
- DOI: 10.1126/sciadv.1601865
Overcoming a nucleosomal barrier to replication
Abstract
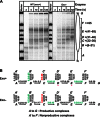
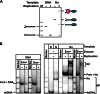
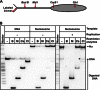
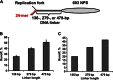
Efficient overcoming and accurate maintenance of chromatin structure and associated histone marks during DNA replication are essential for normal functioning of the daughter cells. However, the molecular mechanisms of replication through chromatin are unknown. We have studied traversal of uniquely positioned mononucleosomes by T7 replisome in vitro. Nucleosomes present a strong, sequence-dependent barrier for replication, with particularly strong pausing of DNA polymerase at the +(31-40) and +(41-65) regions of the nucleosomal DNA. The exonuclease activity of T7 DNA polymerase increases the overall rate of progression of the replisome through a nucleosome, likely by resolving nonproductive complexes. The presence of nucleosome-free DNA upstream of the replication fork facilitates the progression of DNA polymerase through the nucleosome. After replication, at least 50% of the nucleosomes assume an alternative conformation, maintaining their original positions on the DNA. Our data suggest a previously unpublished mechanism for nucleosome maintenance during replication, likely involving transient formation of an intranucleosomal DNA loop.
Keywords: Replication; chromatin; mechanism; nucleosome.
Figures


Similar articles
-
Nucleosome assembly and disassembly activity of GRWD1, a novel Cdt1-binding protein that promotes pre-replication complex formation.Biochim Biophys Acta. 2016 Nov;1863(11):2739-2748. doi: 10.1016/j.bbamcr.2016.08.008. Epub 2016 Aug 20. Biochim Biophys Acta. 2016. PMID: 27552915
-
Coupling of replisome movement with nucleosome dynamics can contribute to the parent-daughter information transfer.Nucleic Acids Res. 2018 Jun 1;46(10):4991-5000. doi: 10.1093/nar/gky207. Nucleic Acids Res. 2018. PMID: 29850895 Free PMC article.
-
Analysis of the mechanism of nucleosome survival during transcription.Nucleic Acids Res. 2014 Feb;42(3):1619-27. doi: 10.1093/nar/gkt1120. Epub 2013 Nov 13. Nucleic Acids Res. 2014. PMID: 24234452 Free PMC article.
-
[Structure and function of histone chaperone FACT].Mol Biol (Mosk). 2015 Nov-Dec;49(6):891-904. doi: 10.7868/S0026898415060026. Mol Biol (Mosk). 2015. PMID: 26710768 Review. Russian.
-
Implications of DNA replication for eukaryotic gene expression.J Cell Sci. 1991 Jun;99 ( Pt 2):201-6. doi: 10.1242/jcs.99.2.201. J Cell Sci. 1991. PMID: 1885667 Review.
Cited by
-
Effective dynamics of nucleosome configurations at the yeast PHO5 promoter.Elife. 2021 Mar 5;10:e58394. doi: 10.7554/eLife.58394. Elife. 2021. PMID: 33666171 Free PMC article.
-
The lane-switch mechanism for nucleosome repositioning by DNA translocase.Nucleic Acids Res. 2021 Sep 20;49(16):9066-9076. doi: 10.1093/nar/gkab664. Nucleic Acids Res. 2021. PMID: 34365508 Free PMC article.
-
Chromatin replication: TRANSmitting the histone code.J Nat Sci. 2017 Feb;3(2):e322. J Nat Sci. 2017. PMID: 28393112 Free PMC article.
-
Rescuing Replication from Barriers: Mechanistic Insights from Single-Molecule Studies.Mol Cell Biol. 2019 Apr 30;39(10):e00576-18. doi: 10.1128/MCB.00576-18. Print 2019 May 15. Mol Cell Biol. 2019. PMID: 30886122 Free PMC article. Review.
-
Structure and function of the histone chaperone FACT - Resolving FACTual issues.Biochim Biophys Acta Gene Regul Mech. 2018 Jul 25:S1874-9399(18)30159-7. doi: 10.1016/j.bbagrm.2018.07.008. Online ahead of print. Biochim Biophys Acta Gene Regul Mech. 2018. PMID: 30055319 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

