RanBPM (RanBP9) regulates mouse c-Kit receptor level and is essential for normal development of bone marrow progenitor cells
- PMID: 27835883
- PMCID: PMC5341297
- DOI: 10.18632/oncotarget.13198
RanBPM (RanBP9) regulates mouse c-Kit receptor level and is essential for normal development of bone marrow progenitor cells
Abstract
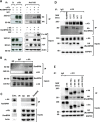
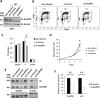
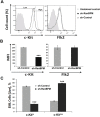
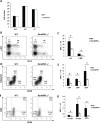
c-Kit is a tyrosine kinase receptor important for gametogenesis, hematopoiesis, melanogenesis and mast cell biology. Dysregulation of c-Kit function is oncogenic and its expression in the stem cell niche of a number of tissues has underlined its relevance for regenerative medicine and hematopoietic stem cell biology. Yet, very little is known about the mechanisms that control c-Kit protein levels. Here we show that the RanBPM/RanBP9 scaffold protein binds to c-Kit and is necessary for normal c-Kit protein expression in the mouse testis and subset lineages of the hematopoietic system. RanBPM deletion causes a reduction in c-Kit protein but not its mRNA suggesting a posttranslational mechanism. This regulation is specific to the c-Kit receptor since RanBPM reduction does not affect other membrane proteins examined. Importantly, in both mouse hematopoietic system and testis, RanBPM deficiency causes defects consistent with c-Kit loss of expression suggesting that RanBPM is an important regulator of c-Kit function. The finding that this regulatory mechanism is also present in human cells expressing endogenous RanBPM and c-Kit suggests a potential new strategy to target oncogenic c-Kit in malignancies.
Keywords: RanBP9; c-Kit signaling; hematopoietic system; spermatogenesis; stem cells.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

Similar articles
-
Ran-binding protein M is associated with human spermatogenesis and oogenesis.Mol Med Rep. 2018 Feb;17(2):2257-2262. doi: 10.3892/mmr.2017.8147. Epub 2017 Nov 23. Mol Med Rep. 2018. PMID: 29207172 Free PMC article.
-
A novel MET-interacting protein shares high sequence similarity with RanBPM, but fails to stimulate MET-induced Ras/Erk signaling.Biochem Biophys Res Commun. 2004 Jan 9;313(2):320-6. doi: 10.1016/j.bbrc.2003.11.124. Biochem Biophys Res Commun. 2004. PMID: 14684163
-
RanBPM, a scaffolding protein for gametogenesis.Curr Top Dev Biol. 2013;102:357-84. doi: 10.1016/B978-0-12-416024-8.00013-1. Curr Top Dev Biol. 2013. PMID: 23287040 Free PMC article. Review.
-
Diverse roles of the scaffolding protein RanBPM.Drug Discov Today. 2012 Apr;17(7-8):379-87. doi: 10.1016/j.drudis.2011.10.030. Epub 2011 Nov 7. Drug Discov Today. 2012. PMID: 22094242 Review.
-
Inhibition of HDAC6 activity through interaction with RanBPM and its associated CTLH complex.BMC Cancer. 2017 Jul 1;17(1):460. doi: 10.1186/s12885-017-3430-2. BMC Cancer. 2017. PMID: 28668087 Free PMC article.
Cited by
-
The CTLH Complex in Cancer Cell Plasticity.J Oncol. 2019 Nov 30;2019:4216750. doi: 10.1155/2019/4216750. eCollection 2019. J Oncol. 2019. PMID: 31885576 Free PMC article. Review.
-
Regulation of normal and leukemic stem cells through cytokine signaling and the microenvironment.Int J Hematol. 2017 May;105(5):566-577. doi: 10.1007/s12185-017-2184-6. Epub 2017 Feb 7. Int J Hematol. 2017. PMID: 28176225 Review.
-
Mechanically strained osteocyte-derived exosomes contained miR-3110-5p and miR-3058-3p and promoted osteoblastic differentiation.Biomed Eng Online. 2024 May 5;23(1):44. doi: 10.1186/s12938-024-01237-9. Biomed Eng Online. 2024. PMID: 38705993 Free PMC article.
-
Cell signalling pathway regulation by RanBPM: molecular insights and disease implications.Open Biol. 2017 Jun;7(6):170081. doi: 10.1098/rsob.170081. Open Biol. 2017. PMID: 28659384 Free PMC article. Review.
-
Bone marrow Tregs mediate stromal cell function and support hematopoiesis via IL-10.JCI Insight. 2020 Nov 19;5(22):e135681. doi: 10.1172/jci.insight.135681. JCI Insight. 2020. PMID: 33208555 Free PMC article.
References
-
- Morrison-Graham K, Takahashi Y. Steel factor and c-kit receptor: from mutants to a growth factor system. BioEssays. 1993;15:77–83. - PubMed
-
- Lennartsson J, Ronnstrand L. Stem cell factor receptor/c-Kit: from basic science to clinical implications. Physiological reviews. 2012;92:1619–1649. - PubMed
-
- Rojas-Sutterlin S, Lecuyer E, Hoang T. Kit and Scl regulation of hematopoietic stem cells. Curr Opin Hematol. 2014;21:256–264. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

