High resolution microscopy reveals the nuclear shape of budding yeast during cell cycle and in various biological states
- PMID: 27831493
- PMCID: PMC5201014
- DOI: 10.1242/jcs.188250
High resolution microscopy reveals the nuclear shape of budding yeast during cell cycle and in various biological states
Abstract
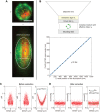
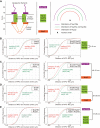
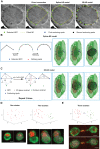
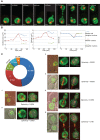
How spatial organization of the genome depends on nuclear shape is unknown, mostly because accurate nuclear size and shape measurement is technically challenging. In large cell populations of the yeast Saccharomyces cerevisiae, we assessed the geometry (size and shape) of nuclei in three dimensions with a resolution of 30 nm. We improved an automated fluorescence localization method by implementing a post-acquisition correction of the spherical microscopic aberration along the z-axis, to detect the three dimensional (3D) positions of nuclear pore complexes (NPCs) in the nuclear envelope. Here, we used a method called NucQuant to accurately estimate the geometry of nuclei in 3D throughout the cell cycle. To increase the robustness of the statistics, we aggregated thousands of detected NPCs from a cell population in a single representation using the nucleolus or the spindle pole body (SPB) as references to align nuclei along the same axis. We could detect asymmetric changes of the nucleus associated with modification of nucleolar size. Stereotypical modification of the nucleus toward the nucleolus further confirmed the asymmetric properties of the nuclear envelope.
Keywords: Localization microscopy; Nuclear geometry; Nuclear pore complex; Super resolution microscopy.
© 2016. Published by The Company of Biologists Ltd.
Conflict of interest statement
The authors declare no competing or financial interests.
Figures


Similar articles
-
Nuclear envelope expansion in budding yeast is independent of cell growth and does not determine nuclear volume.Mol Biol Cell. 2019 Jan 1;30(1):131-145. doi: 10.1091/mbc.E18-04-0204. Epub 2018 Oct 31. Mol Biol Cell. 2019. PMID: 30379612 Free PMC article.
-
The Malleable Nature of the Budding Yeast Nuclear Envelope: Flares, Fusion, and Fenestrations.J Cell Physiol. 2016 Nov;231(11):2353-60. doi: 10.1002/jcp.25355. Epub 2016 Apr 8. J Cell Physiol. 2016. PMID: 26909870 Free PMC article. Review.
-
Nuclear pore complex antigens delineate nuclear envelope dynamics in vegetative and conjugating Saccharomyces cerevisiae.Yeast. 1993 Mar;9(3):235-49. doi: 10.1002/yea.320090304. Yeast. 1993. PMID: 8488725
-
High-Resolution Microscopy to Learn the Nuclear Organization of the Living Yeast Cells.Stem Cells Int. 2021 Aug 27;2021:9951114. doi: 10.1155/2021/9951114. eCollection 2021. Stem Cells Int. 2021. Retraction in: Stem Cells Int. 2024 Jan 24;2024:9872132. doi: 10.1155/2024/9872132 PMID: 34497652 Free PMC article. Retracted.
-
Dynamics of nuclear pore complex organization through the cell cycle.Curr Opin Cell Biol. 2004 Jun;16(3):314-21. doi: 10.1016/j.ceb.2004.04.001. Curr Opin Cell Biol. 2004. PMID: 15145357 Review.
Cited by
-
Raman Spectroscopy-Based Measurements of Single-Cell Phenotypic Diversity in Microbial Populations.mSphere. 2020 Oct 28;5(5):e00806-20. doi: 10.1128/mSphere.00806-20. mSphere. 2020. PMID: 33115836 Free PMC article.
-
Nucleolar localization of the yeast RNA exosome subunit Rrp44 hints at early pre-rRNA processing as its main function.J Biol Chem. 2020 Aug 7;295(32):11195-11213. doi: 10.1074/jbc.RA120.013589. Epub 2020 Jun 17. J Biol Chem. 2020. PMID: 32554806 Free PMC article.
-
Quantitative analysis of nuclear pore complex organization in Schizosaccharomyces pombe.Life Sci Alliance. 2022 Mar 30;5(7):e202201423. doi: 10.26508/lsa.202201423. Print 2022 Jul. Life Sci Alliance. 2022. PMID: 35354597 Free PMC article.
-
Nuclear envelope expansion in budding yeast is independent of cell growth and does not determine nuclear volume.Mol Biol Cell. 2019 Jan 1;30(1):131-145. doi: 10.1091/mbc.E18-04-0204. Epub 2018 Oct 31. Mol Biol Cell. 2019. PMID: 30379612 Free PMC article.
-
The Intracellular Cleavage Product of the NG2 Proteoglycan Modulates Translation and Cell-Cycle Kinetics via Effects on mTORC1/FMRP Signaling.Front Cell Neurosci. 2018 Aug 7;12:231. doi: 10.3389/fncel.2018.00231. eCollection 2018. Front Cell Neurosci. 2018. PMID: 30131676 Free PMC article.
References
-
- Albert B., Léger-Silvestre I., Normand C., Ostermaier M., Pérez-Fernández J., Panov K., Zomerdijk J. C. B. M., Schultz P. and Gadal O. (2011). RNA polymerase I–specific subunits promote polymerase clustering to enhance the rRNA gene transcription cycle. J. Cell Biol. 192, 277-293. 10.1083/jcb.201006040 - DOI - PMC - PubMed
-
- Albert B., Mathon J., Shukla A., Saad H., Normand C., Léger-Silvestre I., Villa D., Kamgoue A., Mozziconacci J., Wong H. et al. (2013). Systematic characterization of the conformation and dynamics of budding yeast chromosome XII. J. Cell Biol. 202, 201-210. 10.1083/jcb.201208186 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

