Gene expression profiles and protein-protein interaction networks in amyotrophic lateral sclerosis patients with C9orf72 mutation
- PMID: 27814735
- PMCID: PMC5097384
- DOI: 10.1186/s13023-016-0531-y
Gene expression profiles and protein-protein interaction networks in amyotrophic lateral sclerosis patients with C9orf72 mutation
Abstract
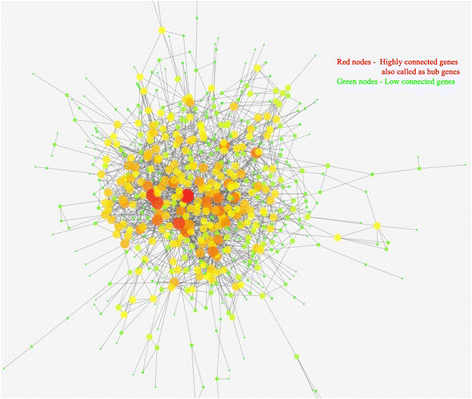
Background: Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disease that involves the death of neurons. ALS is associated with many gene mutations as previously studied. In order to explore the molecular mechanisms underlying ALS with C9orf72 mutation, gene expression profiles of ALS fibroblasts and control fibroblasts were subjected to bioinformatics analysis. Genes with critical functional roles can be detected by a measure of node centrality in biological networks. In gene co-expression networks, highly connected genes called as candidate hubs have been associated with key disease-related pathways. Herein, this method was applied to find the hub genes related to ALS disease.
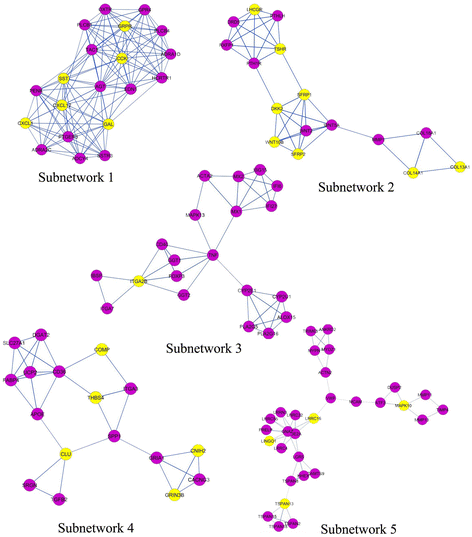
Methods: Illumina HiSeq microarray gene expression dataset GSE51684 was retrieved from Gene Expression Omnibus (GEO) database which included four Sporadic ALS, twelve Familial ALS and eight control samples. Differentially Expressed Genes (DEGs) were identified using the Student's t test statistical method and gene co-expression networking. Gene ontology (GO) function and KEGG pathway enrichment analysis of DEGs were performed using the DAVID online tool. Protein-protein interaction (PPI) networks were constructed by mapping the DEGs onto protein-protein interaction data from publicly available databases to identify the pathways where DEGs are involved in. PPI interaction network was divided into subnetworks using MCODE algorithm and was analyzed using Cytoscape.
Results: The results revealed that the expression of DEGs was mainly involved in cell adhesion, cell-cell signaling, Extra cellular matrix region GO processes and focal adhesion, neuroactive ligand receptor interaction, Extracellular matrix receptor interaction. Tumor necrosis factor (TNF), Endothelin 1 (EDN1), Angiotensin (AGT) and many cell adhesion molecules (CAM) were detected as hub genes that can be targeted as novel therapeutic targets for ALS disease.
Conclusion: These analyses and findings enhance the understanding of ALS pathogenesis and provide references for ALS therapy.
Keywords: Amyotrophic lateral sclerosis; C9orf72 mutation; Hub genes; Protein-protein interaction network.
Figures
Similar articles
-
Screening and identification of key biomarkers associated with amyotrophic lateral sclerosis and depression using bioinformatics.Medicine (Baltimore). 2023 Nov 24;102(47):e36265. doi: 10.1097/MD.0000000000036265. Medicine (Baltimore). 2023. PMID: 38013317 Free PMC article.
-
Decoding genetic and pathophysiological mechanisms in amyotrophic lateral sclerosis and primary lateral sclerosis: A comparative study of differentially expressed genes and implicated pathways in motor neuron disorders.Adv Protein Chem Struct Biol. 2024;141:177-201. doi: 10.1016/bs.apcsb.2023.12.008. Epub 2024 Jun 25. Adv Protein Chem Struct Biol. 2024. PMID: 38960473
-
Bioinformatics Analysis and Identification of Genes and Molecular Pathways Involved in Synovial Inflammation in Rheumatoid Arthritis.Med Sci Monit. 2019 Mar 27;25:2246-2256. doi: 10.12659/MSM.915451. Med Sci Monit. 2019. PMID: 30916045 Free PMC article.
-
Sporadic Amyotrophic Lateral Sclerosis Skeletal Muscle Transcriptome Analysis: A Comprehensive Examination of Differentially Expressed Genes.Biomolecules. 2024 Mar 20;14(3):377. doi: 10.3390/biom14030377. Biomolecules. 2024. PMID: 38540795 Free PMC article. Review.
-
From Environment to Gene Expression: Epigenetic Methylations and One-Carbon Metabolism in Amyotrophic Lateral Sclerosis.Cells. 2024 Jun 3;13(11):967. doi: 10.3390/cells13110967. Cells. 2024. PMID: 38891099 Free PMC article. Review.
Cited by
-
Human ALS/FTD brain organoid slice cultures display distinct early astrocyte and targetable neuronal pathology.Nat Neurosci. 2021 Nov;24(11):1542-1554. doi: 10.1038/s41593-021-00923-4. Epub 2021 Oct 21. Nat Neurosci. 2021. PMID: 34675437 Free PMC article.
-
Promoting Effect and Potential Mechanism of Lactobacillus pentosus LPQ1-Produced Active Compounds on the Secretion of 5-Hydroxytryptophan.Foods. 2022 Dec 2;11(23):3895. doi: 10.3390/foods11233895. Foods. 2022. PMID: 36496703 Free PMC article.
-
Neuroinflammation as a Factor of Neurodegenerative Disease: Thalidomide Analogs as Treatments.Front Cell Dev Biol. 2019 Dec 4;7:313. doi: 10.3389/fcell.2019.00313. eCollection 2019. Front Cell Dev Biol. 2019. PMID: 31867326 Free PMC article. Review.
-
Endothelin-1, over-expressed in SOD1G93A mice, aggravates injury of NSC34-hSOD1G93A cells through complicated molecular mechanism revealed by quantitative proteomics analysis.Front Cell Neurosci. 2022 Dec 1;16:1069617. doi: 10.3389/fncel.2022.1069617. eCollection 2022. Front Cell Neurosci. 2022. PMID: 36531135 Free PMC article.
-
The Anti-Apoptotic Effect of ASC-Exosomes in an In Vitro ALS Model and Their Proteomic Analysis.Cells. 2019 Sep 14;8(9):1087. doi: 10.3390/cells8091087. Cells. 2019. PMID: 31540100 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

