Post-transcriptional gene regulation by mRNA modifications
- PMID: 27808276
- PMCID: PMC5167638
- DOI: 10.1038/nrm.2016.132
Post-transcriptional gene regulation by mRNA modifications
Erratum in
-
Publisher Correction: Post-transcriptional gene regulation by mRNA modifications.Nat Rev Mol Cell Biol. 2018 Dec;19(12):808. doi: 10.1038/s41580-018-0075-1. Nat Rev Mol Cell Biol. 2018. PMID: 30341428
Abstract
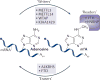
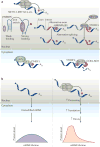
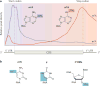
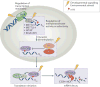
The recent discovery of reversible mRNA methylation has opened a new realm of post-transcriptional gene regulation in eukaryotes. The identification and functional characterization of proteins that specifically recognize RNA N6-methyladenosine (m6A) unveiled it as a modification that cells utilize to accelerate mRNA metabolism and translation. N6-adenosine methylation directs mRNAs to distinct fates by grouping them for differential processing, translation and decay in processes such as cell differentiation, embryonic development and stress responses. Other mRNA modifications, including N1-methyladenosine (m1A), 5-methylcytosine (m5C) and pseudouridine, together with m6A form the epitranscriptome and collectively code a new layer of information that controls protein synthesis.
Conflict of interest statement
statement The authors declare no competing interests.
Figures

Similar articles
-
[Progress in epigenetic modification of mRNA and the function of m6A modification].Sheng Wu Gong Cheng Xue Bao. 2019 May 25;35(5):775-783. doi: 10.13345/j.cjb.180416. Sheng Wu Gong Cheng Xue Bao. 2019. PMID: 31222996 Review. Chinese.
-
Messenger RNA modifications: Form, distribution, and function.Science. 2016 Jun 17;352(6292):1408-12. doi: 10.1126/science.aad8711. Science. 2016. PMID: 27313037 Free PMC article. Review.
-
Translating the epitranscriptome.Wiley Interdiscip Rev RNA. 2017 Jan;8(1):e1375. doi: 10.1002/wrna.1375. Epub 2016 Jun 27. Wiley Interdiscip Rev RNA. 2017. PMID: 27345446 Free PMC article. Review.
-
N6-methyladenosine is required for the hypoxic stabilization of specific mRNAs.RNA. 2017 Sep;23(9):1444-1455. doi: 10.1261/rna.061044.117. Epub 2017 Jun 13. RNA. 2017. PMID: 28611253 Free PMC article.
-
RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation.Genes Dev. 2015 Jul 1;29(13):1343-55. doi: 10.1101/gad.262766.115. Genes Dev. 2015. PMID: 26159994 Free PMC article. Review.
Cited by
-
METTL3-mediated m6A modification regulates cell cycle progression of dental pulp stem cells.Stem Cell Res Ther. 2021 Mar 1;12(1):159. doi: 10.1186/s13287-021-02223-x. Stem Cell Res Ther. 2021. PMID: 33648590 Free PMC article.
-
The N6-methyladenosine Epitranscriptomic Landscape of Lung Adenocarcinoma.Cancer Discov. 2024 Nov 1;14(11):2279-2299. doi: 10.1158/2159-8290.CD-23-1212. Cancer Discov. 2024. PMID: 38922581 Free PMC article.
-
NAMPT is a metabolic checkpoint of IFNγ-producing CD4+ T cells in lupus nephritis.Mol Ther. 2023 Jan 4;31(1):193-210. doi: 10.1016/j.ymthe.2022.09.013. Epub 2022 Sep 22. Mol Ther. 2023. PMID: 36146932 Free PMC article.
-
Immunoprecipitation and Sequencing of Acetylated RNA.Bio Protoc. 2019 Jun 20;9(12):e3278. doi: 10.21769/BioProtoc.3278. eCollection 2019 Jun 20. Bio Protoc. 2019. PMID: 33654795 Free PMC article.
-
Crosstalk between RNA m6A Modification and Non-coding RNA Contributes to Cancer Growth and Progression.Mol Ther Nucleic Acids. 2020 Aug 8;22:62-71. doi: 10.1016/j.omtn.2020.08.004. Online ahead of print. Mol Ther Nucleic Acids. 2020. PMID: 32911345 Free PMC article. Review.
References
-
- Perry RP, Kelley DE. Existence of methylated messenger RNA in mouse L cells. Cell. 1974;1:37–42. References 2 and 3 present the first reports of the existence of m6A in the internal region of mRNA.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

