Evolution and Cryo-electron Microscopy Capsid Structure of a North American Bat Adenovirus and Its Relationship to Other Mastadenoviruses
- PMID: 27807242
- PMCID: PMC5215340
- DOI: 10.1128/JVI.01504-16
Evolution and Cryo-electron Microscopy Capsid Structure of a North American Bat Adenovirus and Its Relationship to Other Mastadenoviruses
Abstract
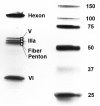
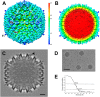
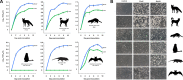
Since the first description of adenoviruses in bats in 2006, a number of micro- and megabat species in Europe, Africa, and Asia have been shown to carry a wide diversity of adenoviruses. Here, we report on the evolutionary, biological, and structural characterization of a novel bat adenovirus (BtAdV) recovered from a Rafinesque's big-eared bat (Corynorhinus rafinesquii) in Kentucky, USA, which is the first adenovirus isolated from North American bats. This virus (BtAdV 250-A) exhibits a close phylogenetic relationship with Canine mastadenovirus A (CAdV A), as previously observed with other BtAdVs. To further investigate the relationships between BtAdVs and CAdVs, we conducted mass spectrometric analysis and single-particle cryo-electron microscopy reconstructions of the BtAdV 250-A capsid and also analyzed the in vitro host ranges of both viruses. Our results demonstrate that BtAdV 250-A represents a new mastadenovirus species that, in contrast to CAdV, has a unique capsid morphology that contains more prominent extensions of protein IX and can replicate efficiently in a phylogenetically diverse range of species. These findings, in addition to the recognition that both the genetic diversity of BtAdVs and the number of different bat species from disparate geographic regions infected with BtAdVs appears to be extensive, tentatively suggest that bats may have served as a potential reservoir for the cross-species transfer of adenoviruses to other hosts, as theorized for CAdV.
Importance: Although many adenoviruses are host specific and likely codiverged with their hosts over millions of years, other adenoviruses appear to have emerged through successful cross-species transmission events on more recent time scales. The wide geographic distribution and genetic diversity of adenoviruses in bats and their close phylogenetic relationship to Canine mastadenovirus A (CAdV A) has raised important questions about how CAdV A, and possibly other mammalian adenoviruses, may have emerged. Although most adenoviruses tend to cause limited disease in their natural hosts, CAdV A is unusual in that it may cause high morbidity and sometimes fatal infections in immunocompetent hosts and is thus an important pathogen of carnivores. Here, we performed a comparative evolutionary and structural study of representative bat and canine adenoviruses to better understand the relationship between these two viral groups.
Keywords: bat adenovirus; canine adenovirus; cross-species transmission; cryo-electron microscopy; host range; mastadenovirus; virus evolution.
Copyright © 2017 American Society for Microbiology.
Figures




Similar articles
-
Characterization of a novel species of adenovirus from Japanese microbat and role of CXADR as its entry factor.Sci Rep. 2019 Jan 24;9(1):573. doi: 10.1038/s41598-018-37224-z. Sci Rep. 2019. PMID: 30679679 Free PMC article.
-
Genetic diversity of adenovirus in neotropical bats from Brazil.Braz J Microbiol. 2023 Dec;54(4):3221-3230. doi: 10.1007/s42770-023-01109-9. Epub 2023 Aug 31. Braz J Microbiol. 2023. PMID: 37653362 Free PMC article.
-
A Novel Mastadenovirus from Nyctalus noctula Which Represents a Distinct Evolutionary Branch of Viruses from Bats in Europe.Viruses. 2024 Jul 26;16(8):1207. doi: 10.3390/v16081207. Viruses. 2024. PMID: 39205181 Free PMC article.
-
Murine adenoviruses: tools for studying adenovirus pathogenesis in a natural host.FEBS Lett. 2019 Dec;593(24):3649-3659. doi: 10.1002/1873-3468.13699. Epub 2019 Dec 6. FEBS Lett. 2019. PMID: 31777948 Free PMC article. Review.
-
Adenoviruses across the animal kingdom: a walk in the zoo.FEBS Lett. 2019 Dec;593(24):3660-3673. doi: 10.1002/1873-3468.13687. Epub 2019 Dec 9. FEBS Lett. 2019. PMID: 31747467 Review.
Cited by
-
Detection and Phylogenetic Characterization of a Novel Adenovirus Found in Lesser Mouse-Eared Bat (Myotis blythii) in South Kazakhstan.Viruses. 2023 May 10;15(5):1139. doi: 10.3390/v15051139. Viruses. 2023. PMID: 37243225 Free PMC article.
-
The Role of Hexon Protein as a Molecular Mold in Patterning the Protein IX Organization in Human Adenoviruses.J Mol Biol. 2017 Sep 1;429(18):2747-2751. doi: 10.1016/j.jmb.2017.06.025. Epub 2017 Jul 18. J Mol Biol. 2017. PMID: 28728980 Free PMC article.
-
Adenovirus Structure: What Is New?Int J Mol Sci. 2021 May 15;22(10):5240. doi: 10.3390/ijms22105240. Int J Mol Sci. 2021. PMID: 34063479 Free PMC article. Review.
-
Characterization of a novel species of adenovirus from Japanese microbat and role of CXADR as its entry factor.Sci Rep. 2019 Jan 24;9(1):573. doi: 10.1038/s41598-018-37224-z. Sci Rep. 2019. PMID: 30679679 Free PMC article.
-
Cryo-EM structure of enteric adenovirus HAdV-F41 highlights structural variations among human adenoviruses.Sci Adv. 2021 Feb 24;7(9):eabd9421. doi: 10.1126/sciadv.abd9421. Print 2021 Feb. Sci Adv. 2021. PMID: 33627423 Free PMC article.
References
-
- Harrach B, Benkö M, Both GW, Brown M, Davison AJ, Echavarría M, Hess M, Jones MS, Kajon A, Lehmkuhl HD, Mautner V, Mittal SK, Wadell G. 2012. Family Adenoviridae, p 125–141. In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA.
-
- Benkö M, Harrach B. 2003. Molecular evolution of adenoviruses. Curr Top Microbiol Immunol 272:3–35. - PubMed
-
- Chen EC, Yagi S, Kelly KR, Mendoza SP, Tarara RP, Canfield DR, Maninger N, Rosenthal A, Spinner A, Bales KL, Schnurr DP, Lerche NW, Chiu CY. 2011. Cross-species transmission of a novel adenovirus associated with a fulminant pneumonia outbreak in a new world monkey colony. PLoS Pathog 7:e1002155. doi:10.1371/journal.ppat.1002155. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

