Large-scale gene co-expression network as a source of functional annotation for cattle genes
- PMID: 27806696
- PMCID: PMC5094014
- DOI: 10.1186/s12864-016-3176-2
Large-scale gene co-expression network as a source of functional annotation for cattle genes
Abstract
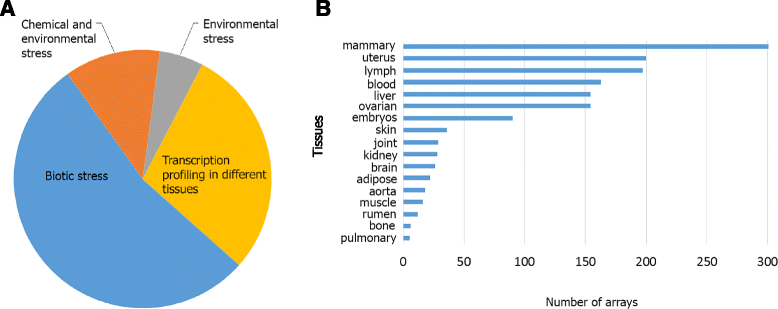
Background: Genome sequencing and subsequent gene annotation of genomes has led to the elucidation of many genes, but in vertebrates the actual number of protein coding genes are very consistent across species (~20,000). Seven years after sequencing the cattle genome, there are still genes that have limited annotation and the function of many genes are still not understood, or partly understood at best. Based on the assumption that genes with similar patterns of expression across a vast array of tissues and experimental conditions are likely to encode proteins with related functions or participate within a given pathway, we constructed a genome-wide Cattle Gene Co-expression Network (CGCN) using 72 microarray datasets that contained a total of 1470 Affymetrix Genechip Bovine Genome Arrays that were retrieved from either NCBI GEO or EBI ArrayExpress.
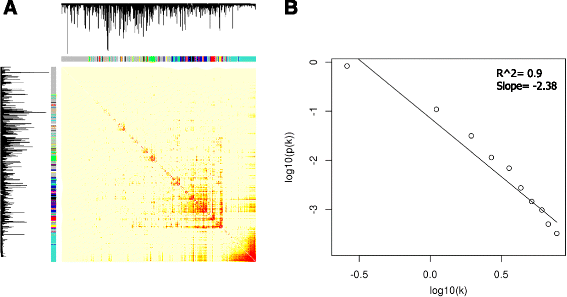
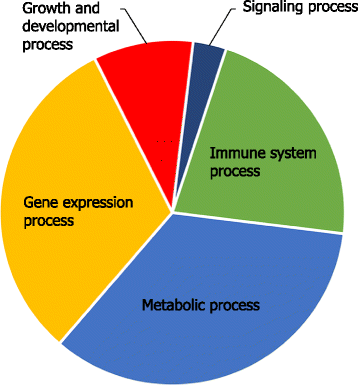
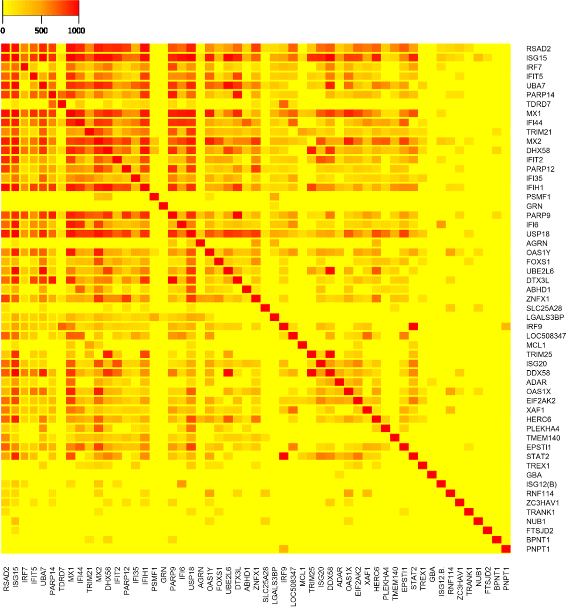
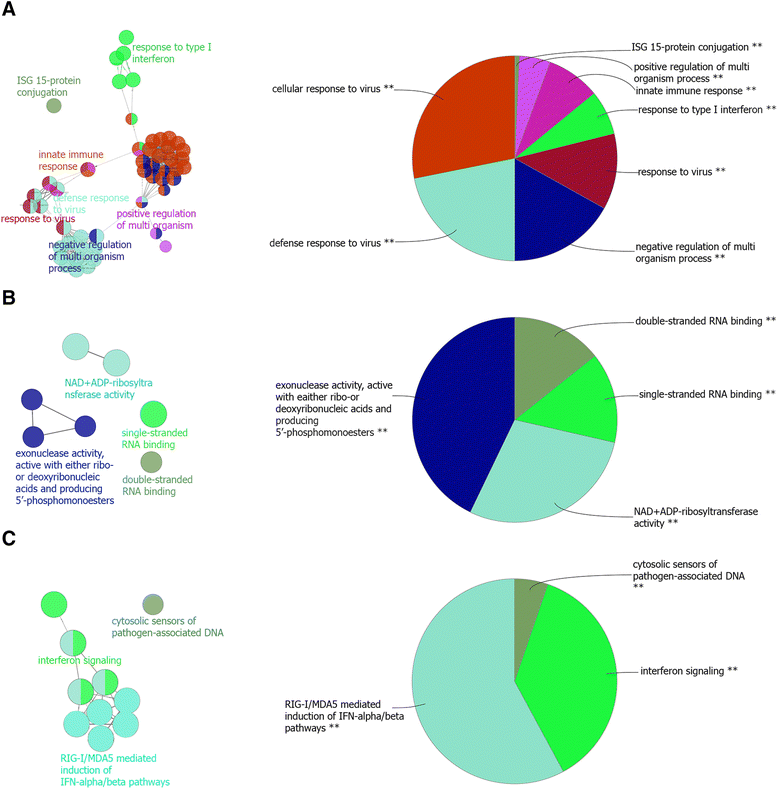
Results: The total of 16,607 probe sets, which represented 11,397 genes, with unique Entrez ID were consolidated into 32 co-expression modules that contained between 29 and 2569 probe sets. All of the identified modules showed strong functional enrichment for gene ontology (GO) terms and Reactome pathways. For example, modules with important biological functions such as response to virus, response to bacteria, energy metabolism, cell signaling and cell cycle have been identified. Moreover, gene co-expression networks using "guilt-by-association" principle have been used to predict the potential function of 132 genes with no functional annotation. Four unknown Hub genes were identified in modules highly enriched for GO terms related to leukocyte activation (LOC509513), RNA processing (LOC100848208), nucleic acid metabolic process (LOC100850151) and organic-acid metabolic process (MGC137211). Such highly connected genes should be investigated more closely as they likely to have key regulatory roles.
Conclusions: We have demonstrated that the CGCN and its corresponding regulons provides rich information for experimental biologists to design experiments, interpret experimental results, and develop novel hypothesis on gene function in this poorly annotated genome. The network is publicly accessible at http://www.animalgenome.org/cgi-bin/host/reecylab/d .
Figures
Similar articles
-
Cattle infection response network and its functional modules.BMC Immunol. 2018 Jan 5;19(1):2. doi: 10.1186/s12865-017-0238-4. BMC Immunol. 2018. PMID: 29301495 Free PMC article.
-
Annotation of gene function in citrus using gene expression information and co-expression networks.BMC Plant Biol. 2014 Jul 15;14:186. doi: 10.1186/1471-2229-14-186. BMC Plant Biol. 2014. PMID: 25023870 Free PMC article.
-
Key regulators in prostate cancer identified by co-expression module analysis.BMC Genomics. 2014 Nov 24;15:1015. doi: 10.1186/1471-2164-15-1015. BMC Genomics. 2014. PMID: 25418933 Free PMC article.
-
Improvements to cardiovascular gene ontology.Atherosclerosis. 2009 Jul;205(1):9-14. doi: 10.1016/j.atherosclerosis.2008.10.014. Epub 2008 Nov 1. Atherosclerosis. 2009. PMID: 19046747 Free PMC article. Review.
-
Computational methods for the identification of differential and coordinated gene expression.Hum Mol Genet. 1999;8(10):1821-32. doi: 10.1093/hmg/8.10.1821. Hum Mol Genet. 1999. PMID: 10469833 Review.
Cited by
-
Cross-population selection signatures in Canchim composite beef cattle.PLoS One. 2022 Apr 1;17(4):e0264279. doi: 10.1371/journal.pone.0264279. eCollection 2022. PLoS One. 2022. PMID: 35363779 Free PMC article.
-
Using Machine Learning to Measure Relatedness Between Genes: A Multi-Features Model.Sci Rep. 2019 Mar 12;9(1):4192. doi: 10.1038/s41598-019-40780-7. Sci Rep. 2019. PMID: 30862804 Free PMC article.
-
Cattle infection response network and its functional modules.BMC Immunol. 2018 Jan 5;19(1):2. doi: 10.1186/s12865-017-0238-4. BMC Immunol. 2018. PMID: 29301495 Free PMC article.
-
Construction, analysis and validation of co-expression network to understand stress adaptation in Deinococcus radiodurans R1.PLoS One. 2020 Jun 24;15(6):e0234721. doi: 10.1371/journal.pone.0234721. eCollection 2020. PLoS One. 2020. PMID: 32579573 Free PMC article.
-
Analysis of protein-protein interaction network based on transcriptome profiling of ovine granulosa cells identifies candidate genes in cyclic recruitment of ovarian follicles.J Anim Sci Technol. 2018 Jun 11;60:11. doi: 10.1186/s40781-018-0171-y. eCollection 2018. J Anim Sci Technol. 2018. PMID: 29992036 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

