A Mouse Model of Chronic West Nile Virus Disease
- PMID: 27806117
- PMCID: PMC5091767
- DOI: 10.1371/journal.ppat.1005996
A Mouse Model of Chronic West Nile Virus Disease
Abstract
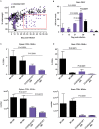
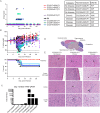
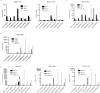
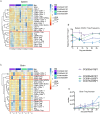
Infection with West Nile virus (WNV) leads to a range of disease outcomes, including chronic infection, though lack of a robust mouse model of chronic WNV infection has precluded identification of the immune events contributing to persistent infection. Using the Collaborative Cross, a population of recombinant inbred mouse strains with high levels of standing genetic variation, we have identified a mouse model of persistent WNV disease, with persistence of viral loads within the brain. Compared to lines exhibiting no disease or marked disease, the F1 cross CC(032x013)F1 displays a strong immunoregulatory signature upon infection that correlates with restraint of the WNV-directed cytolytic response. We hypothesize that this regulatory T cell response sufficiently restrains the immune response such that a chronic infection can be maintained in the CNS. Use of this new mouse model of chronic neuroinvasive virus will be critical in developing improved strategies to prevent prolonged disease in humans.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Immune Correlates of Protection From West Nile Virus Neuroinvasion and Disease.J Infect Dis. 2019 Mar 15;219(7):1162-1171. doi: 10.1093/infdis/jiy623. J Infect Dis. 2019. PMID: 30371803 Free PMC article.
-
Genetic diversity in the collaborative cross model recapitulates human West Nile virus disease outcomes.mBio. 2015 May 5;6(3):e00493-15. doi: 10.1128/mBio.00493-15. mBio. 2015. PMID: 25944860 Free PMC article.
-
Dynamics of Tissue-Specific CD8+ T Cell Responses during West Nile Virus Infection.J Virol. 2018 Apr 27;92(10):e00014-18. doi: 10.1128/JVI.00014-18. Print 2018 May 15. J Virol. 2018. PMID: 29514902 Free PMC article.
-
Immunological headgear: antiviral immune responses protect against neuroinvasive West Nile virus.Trends Mol Med. 2008 Jul;14(7):286-94. doi: 10.1016/j.molmed.2008.05.004. Epub 2008 Jun 6. Trends Mol Med. 2008. PMID: 18539532 Review.
-
Role of Immune Aging in Susceptibility to West Nile Virus.Methods Mol Biol. 2016;1435:235-47. doi: 10.1007/978-1-4939-3670-0_18. Methods Mol Biol. 2016. PMID: 27188562 Free PMC article. Review.
Cited by
-
Immune Predictors of Mortality After Ribonucleic Acid Virus Infection.J Infect Dis. 2020 Mar 2;221(6):882-889. doi: 10.1093/infdis/jiz531. J Infect Dis. 2020. PMID: 31621854 Free PMC article.
-
Giving the Genes a Shuffle: Using Natural Variation to Understand Host Genetic Contributions to Viral Infections.Trends Genet. 2018 Oct;34(10):777-789. doi: 10.1016/j.tig.2018.07.005. Epub 2018 Aug 18. Trends Genet. 2018. PMID: 30131185 Free PMC article. Review.
-
Genetic Diversity of Collaborative Cross Mice Controls Viral Replication, Clinical Severity, and Brain Pathology Induced by Zika Virus Infection, Independently of Oas1b.J Virol. 2020 Jan 17;94(3):e01034-19. doi: 10.1128/JVI.01034-19. Print 2020 Jan 17. J Virol. 2020. PMID: 31694939 Free PMC article.
-
Diverse CD8 T Cell Responses to Viral Infection Revealed by the Collaborative Cross.Cell Rep. 2020 Apr 14;31(2):107508. doi: 10.1016/j.celrep.2020.03.072. Cell Rep. 2020. PMID: 32294433 Free PMC article.
-
Baseline T cell immune phenotypes predict virologic and disease control upon SARS-CoV infection in Collaborative Cross mice.PLoS Pathog. 2021 Jan 29;17(1):e1009287. doi: 10.1371/journal.ppat.1009287. eCollection 2021 Jan. PLoS Pathog. 2021. PMID: 33513210 Free PMC article.
References
-
- Sejvar JJ (2007) The long-term outcomes of human West Nile virus infection. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America 44: 1617–1624. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

