Second generation noninvasive fetal genome analysis reveals de novo mutations, single-base parental inheritance, and preferred DNA ends
- PMID: 27799561
- PMCID: PMC5167168
- DOI: 10.1073/pnas.1615800113
Second generation noninvasive fetal genome analysis reveals de novo mutations, single-base parental inheritance, and preferred DNA ends
Abstract
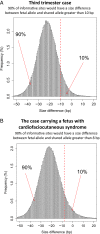
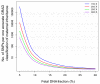
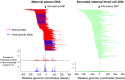
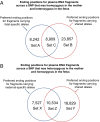
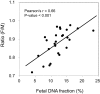
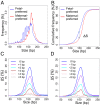
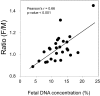
Plasma DNA obtained from a pregnant woman was sequenced to a depth of 270× haploid genome coverage. Comparing the maternal plasma DNA sequencing data with the parental genomic DNA data and using a series of bioinformatics filters, fetal de novo mutations were detected at a sensitivity of 85% and a positive predictive value of 74%. These results represent a 169-fold improvement in the positive predictive value over previous attempts. Improvements in the interpretation of the sequence information of every base position in the genome allowed us to interrogate the maternal inheritance of the fetus for 618,271 of 656,676 (94.2%) heterozygous SNPs within the maternal genome. The fetal genotype at each of these sites was deduced individually, unlike previously, where the inheritance was determined for a collection of sites within a haplotype. These results represent a 90-fold enhancement in the resolution in determining the fetus's maternal inheritance. Selected genomic locations were more likely to be found at the ends of plasma DNA molecules. We found that a subset of such preferred ends exhibited selectivity for fetal- or maternal-derived DNA in maternal plasma. The ratio of the number of maternal plasma DNA molecules with fetal preferred ends to those with maternal preferred ends showed a correlation with the fetal DNA fraction. Finally, this second generation approach for noninvasive fetal whole-genome analysis was validated in a pregnancy diagnosed with cardiofaciocutaneous syndrome with maternal plasma DNA sequenced to 195× coverage. The causative de novo BRAF mutation was successfully detected through the maternal plasma DNA analysis.
Keywords: DNA fragmentation patterns; massively parallel sequencing; noninvasive prenatal testing.
Conflict of interest statement
R.W.K.C. and Y.M.D.L. received research support from Sequenom, Inc. R.W.K.C. and Y.M.D.L. were consultants to Sequenom, Inc. K.C.A.C., R.W.K.C., and Y.M.D.L. hold equities in Sequenom, Inc. K.C.A.C., R.W.K.C., and Y.M.D.L. are founders of Xcelom and Cirina. K.C.A.C., P.J., and R.W.K.C. are consultants to Xcelom. P.J. is a consultant to Cirina. K.C.A.C., P.J., R.W.K.C., and Y.M.D.L. have filed patent applications (PCT/CN2016/073753 and PCT/CN2016/091531) based on the data generated from this work, which have been licensed to Cirina.
Figures




Comment in
-
Noninvasive prenatal testing to analyze the fetal genome.Proc Natl Acad Sci U S A. 2016 Dec 13;113(50):14173-14175. doi: 10.1073/pnas.1617112113. Epub 2016 Nov 30. Proc Natl Acad Sci U S A. 2016. PMID: 27911833 Free PMC article. No abstract available.
Similar articles
-
Haplotype-based approach for noninvasive prenatal diagnosis of congenital adrenal hyperplasia by maternal plasma DNA sequencing.Gene. 2014 Jul 10;544(2):252-8. doi: 10.1016/j.gene.2014.04.055. Epub 2014 Apr 24. Gene. 2014. PMID: 24768736
-
Noninvasive whole-genome sequencing of a human fetus.Sci Transl Med. 2012 Jun 6;4(137):137ra76. doi: 10.1126/scitranslmed.3004323. Sci Transl Med. 2012. PMID: 22674554 Free PMC article.
-
Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus.Sci Transl Med. 2010 Dec 8;2(61):61ra91. doi: 10.1126/scitranslmed.3001720. Sci Transl Med. 2010. PMID: 21148127
-
Bioinformatics Approaches for Fetal DNA Fraction Estimation in Noninvasive Prenatal Testing.Int J Mol Sci. 2017 Feb 20;18(2):453. doi: 10.3390/ijms18020453. Int J Mol Sci. 2017. PMID: 28230760 Free PMC article. Review.
-
Prenatal Diagnosis Innovation: Genome Sequencing of Maternal Plasma.Annu Rev Med. 2016;67:419-32. doi: 10.1146/annurev-med-091014-115715. Epub 2015 Oct 15. Annu Rev Med. 2016. PMID: 26473414 Review.
Cited by
-
Liquid Biopsy in Neuropsychiatric Disorders: A Step Closer to Precision Medicine.Mol Neurobiol. 2024 Sep 19. doi: 10.1007/s12035-024-04492-y. Online ahead of print. Mol Neurobiol. 2024. PMID: 39298102 Review.
-
Cell-free nucleic acid fragmentomics: A non-invasive window into cellular epigenomes.Transl Oncol. 2024 Nov;49:102085. doi: 10.1016/j.tranon.2024.102085. Epub 2024 Aug 22. Transl Oncol. 2024. PMID: 39178576 Free PMC article. Review.
-
DNA methylation and gene expression as determinants of genome-wide cell-free DNA fragmentation.Nat Commun. 2024 Aug 6;15(1):6690. doi: 10.1038/s41467-024-50850-8. Nat Commun. 2024. PMID: 39107309 Free PMC article.
-
Leveraging cfDNA fragmentomic features in a stacked ensemble model for early detection of esophageal squamous cell carcinoma.Cell Rep Med. 2024 Aug 20;5(8):101664. doi: 10.1016/j.xcrm.2024.101664. Epub 2024 Jul 31. Cell Rep Med. 2024. PMID: 39089259 Free PMC article.
-
Longitudinal integrative cell-free DNA analysis in gestational diabetes mellitus.Cell Rep Med. 2024 Aug 20;5(8):101660. doi: 10.1016/j.xcrm.2024.101660. Epub 2024 Jul 25. Cell Rep Med. 2024. PMID: 39059385 Free PMC article.
References
-
- Lo YM, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350(9076):485–487. - PubMed
-
- Bianchi DW, et al. CARE Study Group DNA sequencing versus standard prenatal aneuploidy screening. N Engl J Med. 2014;370(9):799–808. - PubMed
-
- Norton ME, et al. Cell-free DNA analysis for noninvasive examination of trisomy. N Engl J Med. 2015;372(17):1589–1597. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

