Cryo-EM structure of a CD4-bound open HIV-1 envelope trimer reveals structural rearrangements of the gp120 V1V2 loop
- PMID: 27799557
- PMCID: PMC5135367
- DOI: 10.1073/pnas.1615939113
Cryo-EM structure of a CD4-bound open HIV-1 envelope trimer reveals structural rearrangements of the gp120 V1V2 loop
Abstract
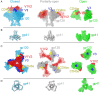
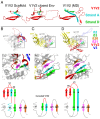
The HIV-1 envelope (Env) glycoprotein, a trimer of gp120-gp41 heterodimers, relies on conformational flexibility to function in fusing the viral and host membranes. Fusion is achieved after gp120 binds to CD4, the HIV-1 receptor, and a coreceptor, capturing an open conformational state in which the fusion machinery on gp41 gains access to the target cell membrane. In the well-characterized closed Env conformation, the gp120 V1V2 loops interact at the apex of the Env trimer. Less is known about the structure of the open CD4-bound state, in which the V1V2 loops must rearrange and separate to allow access to the coreceptor binding site. We identified two anti-HIV-1 antibodies, the coreceptor mimicking antibody 17b and the gp120-gp41 interface-spanning antibody 8ANC195, that can be added as Fabs to a soluble native-like Env trimer to stabilize it in a CD4-bound conformation. Here, we present an 8.9-Å cryo-electron microscopy structure of a BG505 Env-sCD4-17b-8ANC195 complex, which reveals large structural rearrangements in gp120, but small changes in gp41, compared with closed Env structures. The gp120 protomers are rotated and separated in the CD4-bound structure, and the three V1V2 loops are displaced by ∼40 Å from their positions at the trimer apex in closed Env to the sides of the trimer in positions adjacent to, and interacting with, the three bound CD4s. These results are relevant to understanding CD4-induced conformational changes leading to coreceptor binding and fusion, and HIV-1 Env conformational dynamics, and describe a target structure relevant to drug design and vaccine efforts.
Keywords: CD4; HIV-1 Env trimer; HIV-1 coreceptor; conformational change; cryo-EM.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
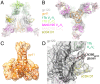

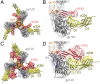
Similar articles
-
Partially Open HIV-1 Envelope Structures Exhibit Conformational Changes Relevant for Coreceptor Binding and Fusion.Cell Host Microbe. 2018 Oct 10;24(4):579-592.e4. doi: 10.1016/j.chom.2018.09.003. Cell Host Microbe. 2018. PMID: 30308160 Free PMC article.
-
Cryo-EM structures of HIV-1 trimer bound to CD4-mimetics BNM-III-170 and M48U1 adopt a CD4-bound open conformation.Nat Commun. 2021 Mar 29;12(1):1950. doi: 10.1038/s41467-021-21816-x. Nat Commun. 2021. PMID: 33782388 Free PMC article.
-
Asymmetric opening of HIV-1 Env bound to CD4 and a coreceptor-mimicking antibody.Nat Struct Mol Biol. 2019 Dec;26(12):1167-1175. doi: 10.1038/s41594-019-0344-5. Epub 2019 Dec 2. Nat Struct Mol Biol. 2019. PMID: 31792452 Free PMC article.
-
Quaternary Interaction of the HIV-1 Envelope Trimer with CD4 and Neutralizing Antibodies.Viruses. 2021 Jul 20;13(7):1405. doi: 10.3390/v13071405. Viruses. 2021. PMID: 34372611 Free PMC article. Review.
-
HIV-1 envelope glycoprotein structure.Curr Opin Struct Biol. 2013 Apr;23(2):268-76. doi: 10.1016/j.sbi.2013.03.007. Epub 2013 Apr 18. Curr Opin Struct Biol. 2013. PMID: 23602427 Free PMC article. Review.
Cited by
-
Electron tomography visualization of HIV-1 fusion with target cells using fusion inhibitors to trap the pre-hairpin intermediate.Elife. 2020 Jul 22;9:e58411. doi: 10.7554/eLife.58411. Elife. 2020. PMID: 32697193 Free PMC article.
-
Antibody Recognition of CD4-Induced Open HIV-1 Env Trimers.J Virol. 2022 Dec 21;96(24):e0108222. doi: 10.1128/jvi.01082-22. Epub 2022 Nov 30. J Virol. 2022. PMID: 36448805 Free PMC article.
-
HIV-1 Env trimers asymmetrically engage CD4 receptors in membranes.Nature. 2023 Nov;623(7989):1026-1033. doi: 10.1038/s41586-023-06762-6. Epub 2023 Nov 22. Nature. 2023. PMID: 37993716 Free PMC article.
-
Global Increases in Human Immunodeficiency Virus Neutralization Sensitivity Due to Alterations in the Membrane-Proximal External Region of the Envelope Glycoprotein Can Be Minimized by Distant State 1-Stabilizing Changes.J Virol. 2022 Apr 13;96(7):e0187821. doi: 10.1128/jvi.01878-21. Epub 2022 Mar 15. J Virol. 2022. PMID: 35289647 Free PMC article.
-
Development of Protein- and Peptide-Based HIV Entry Inhibitors Targeting gp120 or gp41.Viruses. 2019 Aug 1;11(8):705. doi: 10.3390/v11080705. Viruses. 2019. PMID: 31374953 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

