Identification of Novel Kaposi's Sarcoma-Associated Herpesvirus Orf50 Transcripts: Discovery of New RTA Isoforms with Variable Transactivation Potential
- PMID: 27795414
- PMCID: PMC5165194
- DOI: 10.1128/JVI.01434-16
Identification of Novel Kaposi's Sarcoma-Associated Herpesvirus Orf50 Transcripts: Discovery of New RTA Isoforms with Variable Transactivation Potential
Abstract
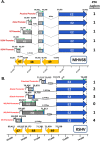
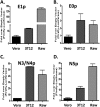
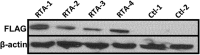
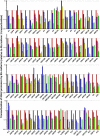
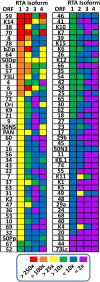
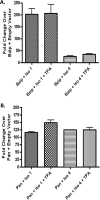
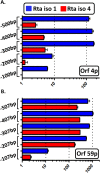
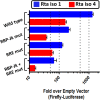
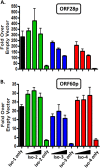
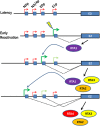
Kaposi's sarcoma-associated herpesvirus (KSHV) is a gammaherpesvirus that has been associated with primary effusion lymphoma and multicentric Castleman's disease, as well as its namesake Kaposi's sarcoma. As a gammaherpesvirus, KSHV is able to acutely replicate, enter latency, and reactivate from this latent state. A key protein involved in both acute replication and reactivation from latency is the replication and transcriptional activator (RTA) encoded by the gene Orf50 RTA is a known transactivator of multiple viral genes, allowing it to control the switch between latency and virus replication. We report here the identification of six alternatively spliced Orf50 transcripts that are generated from four distinct promoters. These newly identified promoters are shown to be transcriptionally active in 293T (embryonic kidney), Vero (African-green monkey kidney epithelial), 3T12 (mouse fibroblast), and RAW 264.7 (mouse macrophage) cell lines. Notably, the newly identified Orf50 transcripts are predicted to encode four different isoforms of the RTA which differ by 6 to 10 residues at the amino terminus of the protein. We show the global viral transactivation potential of all four RTA isoforms and demonstrate that all isoforms can transcriptionally activate an array of KSHV promoters to various levels. The pattern of transcriptional activation appears to support a transcriptional interference model within the Orf50 region, where silencing of previously expressed isoforms by transcription initiation from upstream Orf50 promoters has the potential to modulate the pattern of viral gene activation.
Importance: Gammaherpesviruses are associated with the development of lymphomas and lymphoproliferative diseases, as well as several other types of cancer. The human gammaherpesvirus, Kaposi's sarcoma-associated herpesvirus (KSHV), is tightly associated with the development of Kaposi's sarcoma and multicentric Castleman's disease, as well as a rare form of B cell lymphoma (primary effusion lymphoma) primarily observed in HIV-infected individuals. RTA is an essential viral gene product involved in the initiation of gammaherpesvirus replication and is conserved among all known gammaherpesviruses. We show here for KSHV that transcription of the gene encoding RTA is complex and leads to the expression of several isoforms of RTA with distinct functions. This observed complexity in KSHV RTA expression and function likely plays a critical role in the regulation of downstream viral and cellular gene expression, leading to the efficient production of mature virions.
Keywords: Kaposi's sarcoma-associated herpesvirus; RTA isoforms; alternative promoter usage; alternative splicing; transcriptional activation.
Copyright © 2016 American Society for Microbiology.
Figures

Similar articles
-
Sirtuin 6 Attenuates Kaposi's Sarcoma-Associated Herpesvirus Reactivation by Suppressing Ori-Lyt Activity and Expression of RTA.J Virol. 2019 Mar 21;93(7):e02200-18. doi: 10.1128/JVI.02200-18. Print 2019 Apr 1. J Virol. 2019. PMID: 30651359 Free PMC article.
-
Full-Length Isoforms of Kaposi's Sarcoma-Associated Herpesvirus Latency-Associated Nuclear Antigen Accumulate in the Cytoplasm of Cells Undergoing the Lytic Cycle of Replication.J Virol. 2017 Nov 30;91(24):e01532-17. doi: 10.1128/JVI.01532-17. Print 2017 Dec 15. J Virol. 2017. PMID: 28978712 Free PMC article.
-
Sp3 Transcription Factor Cooperates with the Kaposi's Sarcoma-Associated Herpesvirus ORF50 Protein To Synergistically Activate Specific Viral and Cellular Gene Promoters.J Virol. 2020 Aug 31;94(18):e01143-20. doi: 10.1128/JVI.01143-20. Print 2020 Aug 31. J Virol. 2020. PMID: 32641483 Free PMC article.
-
The Rta/Orf50 transactivator proteins of the gamma-herpesviridae.Curr Top Microbiol Immunol. 2007;312:71-100. doi: 10.1007/978-3-540-34344-8_3. Curr Top Microbiol Immunol. 2007. PMID: 17089794 Review.
-
Epigenetic regulation of Kaposi's sarcoma-associated herpesvirus replication.Semin Cancer Biol. 2009 Jun;19(3):153-7. doi: 10.1016/j.semcancer.2009.02.010. Epub 2009 Feb 21. Semin Cancer Biol. 2009. PMID: 19429478 Free PMC article. Review.
Cited by
-
Autophagy in Viral Development and Progression of Cancer.Front Oncol. 2021 Mar 8;11:603224. doi: 10.3389/fonc.2021.603224. eCollection 2021. Front Oncol. 2021. PMID: 33763351 Free PMC article. Review.
-
Genome-wide regulation of KSHV RNA splicing by viral RNA-binding protein ORF57.PLoS Pathog. 2022 Jul 14;18(7):e1010311. doi: 10.1371/journal.ppat.1010311. eCollection 2022 Jul. PLoS Pathog. 2022. PMID: 35834586 Free PMC article.
-
Kaposi's Sarcoma-Associated Herpesvirus Reactivation by Targeting of a dCas9-Based Transcription Activator to the ORF50 Promoter.Viruses. 2020 Aug 27;12(9):952. doi: 10.3390/v12090952. Viruses. 2020. PMID: 32867368 Free PMC article.
-
Viral Oncology: Molecular Biology and Pathogenesis.J Clin Med. 2017 Nov 29;6(12):111. doi: 10.3390/jcm6120111. J Clin Med. 2017. PMID: 29186062 Free PMC article. Review.
-
Arachidonic Acid Derived Lipid Mediators Influence Kaposi's Sarcoma-Associated Herpesvirus Infection and Pathogenesis.Front Microbiol. 2019 Mar 12;10:358. doi: 10.3389/fmicb.2019.00358. eCollection 2019. Front Microbiol. 2019. PMID: 30915039 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

