Application of RNAi-induced gene expression profiles for prognostic prediction in breast cancer
- PMID: 27788678
- PMCID: PMC5084341
- DOI: 10.1186/s13073-016-0363-3
Application of RNAi-induced gene expression profiles for prognostic prediction in breast cancer
Abstract
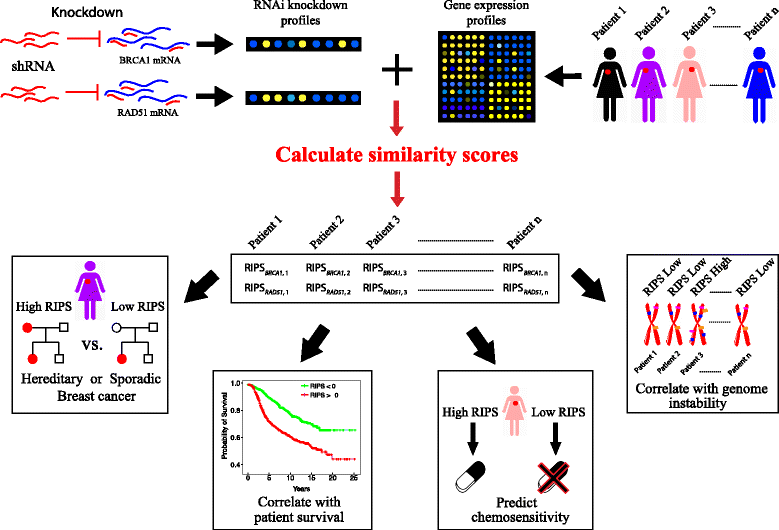
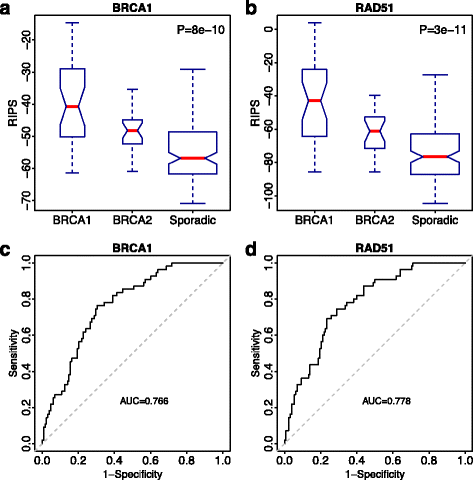
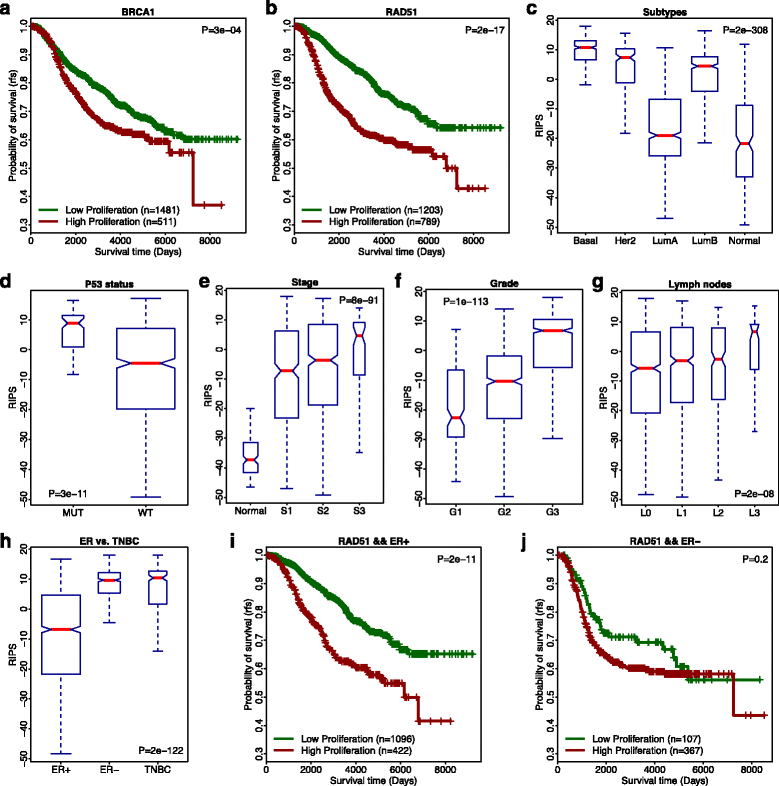
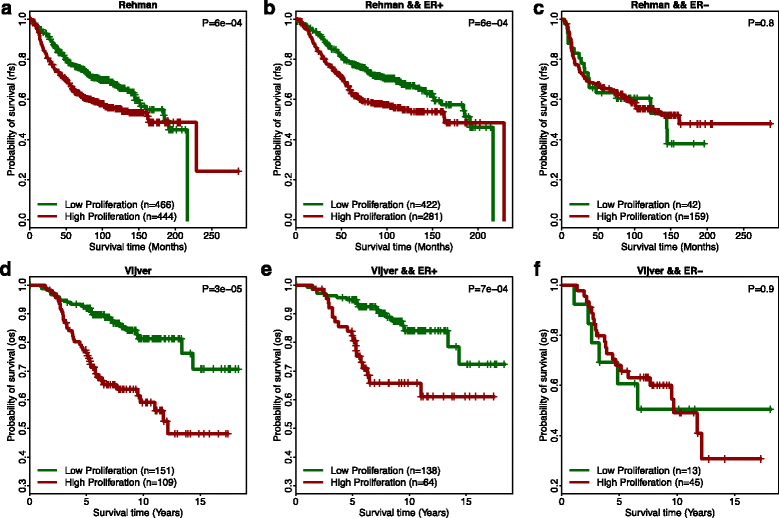
Homologous recombination (HR) is the primary pathway for repairing double-strand DNA breaks implicating in the development of cancer. RNAi-based knockdowns of BRCA1 and RAD51 in this pathway have been performed to investigate the resulting transcriptomic profiles. Here we propose a computational framework to utilize these profiles to calculate a score, named RNA-Interference derived Proliferation Score (RIPS), which reflects cell proliferation ability in individual breast tumors. RIPS is predictive of breast cancer classes, prognosis, genome instability, and neoadjuvant chemosensitivity. This framework directly translates the readout of knockdown experiments into potential clinical applications and generates a robust biomarker in breast cancer.
Keywords: Cancer prognosis; Cell proliferation; Gene knockdown profiles; Genomic instability; Homologous recombination pathway; Neoadjuvant chemotherapy.
Figures
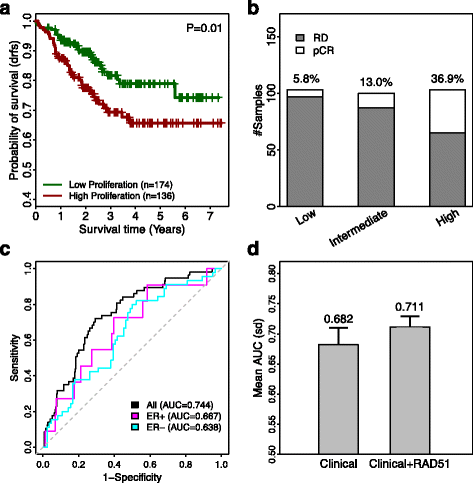
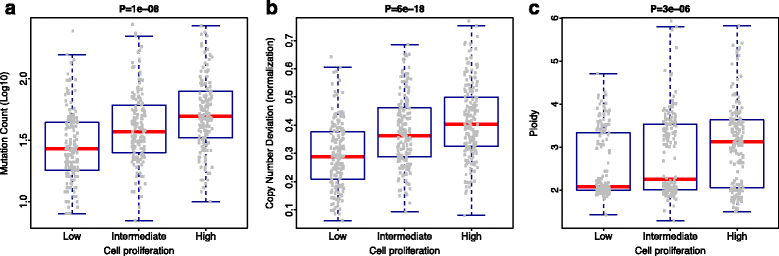
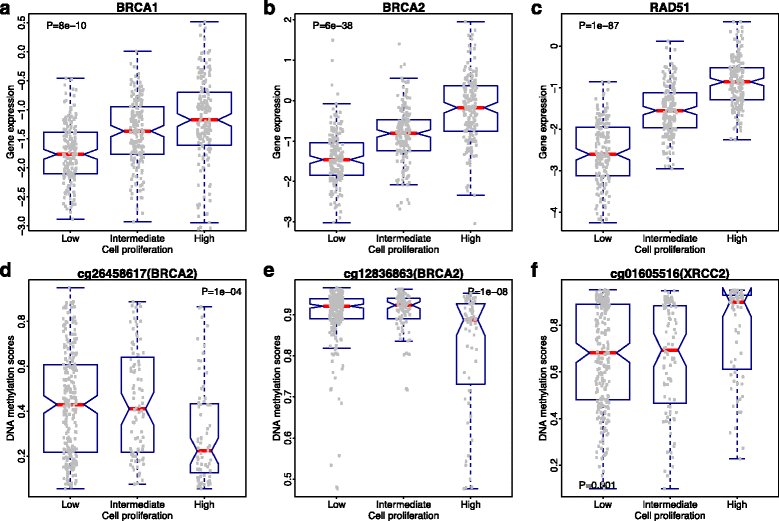
Similar articles
-
Computational Investigation of Homologous Recombination DNA Repair Deficiency in Sporadic Breast Cancer.Sci Rep. 2017 Nov 16;7(1):15742. doi: 10.1038/s41598-017-16138-2. Sci Rep. 2017. PMID: 29146938 Free PMC article.
-
BRCA1 regulates RAD51 function in response to DNA damage and suppresses spontaneous sister chromatid replication slippage: implications for sister chromatid cohesion, genome stability, and carcinogenesis.Cancer Res. 2005 Dec 15;65(24):11384-91. doi: 10.1158/0008-5472.CAN-05-2156. Cancer Res. 2005. PMID: 16357146
-
RAD51 up-regulation bypasses BRCA1 function and is a common feature of BRCA1-deficient breast tumors.Cancer Res. 2007 Oct 15;67(20):9658-65. doi: 10.1158/0008-5472.CAN-07-0290. Cancer Res. 2007. PMID: 17942895
-
Minding the gap: the underground functions of BRCA1 and BRCA2 at stalled replication forks.DNA Repair (Amst). 2007 Jul 1;6(7):1018-31. doi: 10.1016/j.dnarep.2007.02.020. Epub 2007 Mar 26. DNA Repair (Amst). 2007. PMID: 17379580 Free PMC article. Review.
-
Homologous repair of DNA damage and tumorigenesis: the BRCA connection.Oncogene. 2002 Dec 16;21(58):8981-93. doi: 10.1038/sj.onc.1206176. Oncogene. 2002. PMID: 12483514 Review.
Cited by
-
Computational Investigation of Homologous Recombination DNA Repair Deficiency in Sporadic Breast Cancer.Sci Rep. 2017 Nov 16;7(1):15742. doi: 10.1038/s41598-017-16138-2. Sci Rep. 2017. PMID: 29146938 Free PMC article.
-
Cloud bursting galaxy: federated identity and access management.Bioinformatics. 2020 Jan 1;36(1):1-9. doi: 10.1093/bioinformatics/btz472. Bioinformatics. 2020. PMID: 31197310 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

