snRNA 3' End Processing by a CPSF73-Containing Complex Essential for Development in Arabidopsis
- PMID: 27780203
- PMCID: PMC5079582
- DOI: 10.1371/journal.pbio.1002571
snRNA 3' End Processing by a CPSF73-Containing Complex Essential for Development in Arabidopsis
Abstract
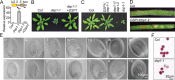
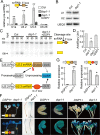
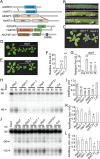
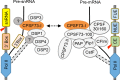
Uridine-rich small nuclear RNAs (snRNAs) are the basal components of the spliceosome and play essential roles in splicing. The biogenesis of the majority of snRNAs involves 3' end endonucleolytic cleavage of the nascent transcript from the elongating DNA-dependent RNA ploymerase II. However, the protein factors responsible for this process remain elusive in plants. Here, we show that DEFECTIVE in snRNA PROCESSING 1 (DSP1) is an essential protein for snRNA 3' end maturation in Arabidopsis. A hypomorphic dsp1-1 mutation causes pleiotropic developmental defects, impairs the 3' end processing of snRNAs, increases the levels of snRNA primary transcripts (pre-snRNAs), and alters the occupancy of Pol II at snRNA loci. In addition, DSP1 binds snRNA loci and interacts with Pol-II in a DNA/RNA-dependent manner. We further show that DSP1 forms a conserved complex, which contains at least four additional proteins, to catalyze snRNA 3' end maturation in Arabidopsis. The catalytic component of this complex is likely the cleavage and polyadenylation specificity factor 73 kDa-I (CSPF73-I), which is the nuclease cleaving the pre-mRNA 3' end. However, the DSP1 complex does not affect pre-mRNA 3' end cleavage, suggesting that plants may use different CPSF73-I-containing complexes to process snRNAs and pre-mRNAs. This study identifies a complex responsible for the snRNA 3' end maturation in plants and uncovers a previously unknown function of CPSF73 in snRNA maturation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Alternative splicing of DSP1 enhances snRNA accumulation by promoting transcription termination and recycle of the processing complex.Proc Natl Acad Sci U S A. 2020 Aug 18;117(33):20325-20333. doi: 10.1073/pnas.2002115117. Epub 2020 Aug 3. Proc Natl Acad Sci U S A. 2020. PMID: 32747542 Free PMC article.
-
DSP1 and DSP4 Act Synergistically in Small Nuclear RNA 3' End Maturation and Pollen Growth.Plant Physiol. 2019 Aug;180(4):2142-2151. doi: 10.1104/pp.19.00231. Epub 2019 Jun 21. Plant Physiol. 2019. PMID: 31227618 Free PMC article.
-
Activity of chimeric U small nuclear RNA (snRNA)/mRNA genes in transfected protoplasts of Nicotiana plumbaginifolia: U snRNA 3'-end formation and transcription initiation can occur independently in plants.Mol Cell Biol. 1993 Oct;13(10):6403-15. doi: 10.1128/mcb.13.10.6403-6415.1993. Mol Cell Biol. 1993. PMID: 8413239 Free PMC article.
-
Transcriptional regulation of snRNAs and its significance for plant development.J Plant Res. 2017 Jan;130(1):57-66. doi: 10.1007/s10265-016-0883-3. Epub 2016 Nov 29. J Plant Res. 2017. PMID: 27900551 Review.
-
Spliceosomal snRNAs, the Essential Players in pre-mRNA Processing in Eukaryotic Nucleus: From Biogenesis to Functions and Spatiotemporal Characteristics.Adv Biol (Weinh). 2024 Jul;8(7):e2400006. doi: 10.1002/adbi.202400006. Epub 2024 May 26. Adv Biol (Weinh). 2024. PMID: 38797893 Review.
Cited by
-
Natural variation in the plant polyadenylation complex.Front Plant Sci. 2024 Jan 22;14:1303398. doi: 10.3389/fpls.2023.1303398. eCollection 2023. Front Plant Sci. 2024. PMID: 38317838 Free PMC article.
-
KinG Is a Plant-Specific Kinesin That Regulates Both Intra- and Intercellular Movement of SHORT-ROOT.Plant Physiol. 2018 Jan;176(1):392-405. doi: 10.1104/pp.17.01518. Epub 2017 Nov 9. Plant Physiol. 2018. PMID: 29122988 Free PMC article.
-
Genomic regulation of transcription and RNA processing by the multitasking Integrator complex.Nat Rev Mol Cell Biol. 2023 Mar;24(3):204-220. doi: 10.1038/s41580-022-00534-2. Epub 2022 Sep 30. Nat Rev Mol Cell Biol. 2023. PMID: 36180603 Free PMC article. Review.
-
Alternative splicing of DSP1 enhances snRNA accumulation by promoting transcription termination and recycle of the processing complex.Proc Natl Acad Sci U S A. 2020 Aug 18;117(33):20325-20333. doi: 10.1073/pnas.2002115117. Epub 2020 Aug 3. Proc Natl Acad Sci U S A. 2020. PMID: 32747542 Free PMC article.
-
DNA-dependent RNA polymerases in plants.Plant Cell. 2023 Sep 27;35(10):3641-3661. doi: 10.1093/plcell/koad195. Plant Cell. 2023. PMID: 37453082 Free PMC article.
References
-
- Carbon P, Murgo S, Ebel JP, Krol A, Tebb G, Mattaj LW. A common octamer motif binding protein is involved in the transcription of U6 snRNA by RNA polymerase III and U2 snRNA by RNA polymerase II. Cell. 1987;51(1):71–9. . - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

