A Proteogenomic Approach to Understanding MYC Function in Metastatic Medulloblastoma Tumors
- PMID: 27775567
- PMCID: PMC5085772
- DOI: 10.3390/ijms17101744
A Proteogenomic Approach to Understanding MYC Function in Metastatic Medulloblastoma Tumors
Abstract
Brain tumors are the leading cause of cancer-related deaths in children, and medulloblastoma is the most prevalent malignant childhood/pediatric brain tumor. Providing effective treatment for these cancers, with minimal damage to the still-developing brain, remains one of the greatest challenges faced by clinicians. Understanding the diverse events driving tumor formation, maintenance, progression, and recurrence is necessary for identifying novel targeted therapeutics and improving survival of patients with this disease. Genomic copy number alteration data, together with clinical studies, identifies c-MYC amplification as an important risk factor associated with the most aggressive forms of medulloblastoma with marked metastatic potential. Yet despite this, very little is known regarding the impact of such genomic abnormalities upon the functional biology of the tumor cell. We discuss here how recent advances in quantitative proteomic techniques are now providing new insights into the functional biology of these aggressive tumors, as illustrated by the use of proteomics to bridge the gap between the genotype and phenotype in the case of c-MYC-amplified/associated medulloblastoma. These integrated proteogenomic approaches now provide a new platform for understanding cancer biology by providing a functional context to frame genomic abnormalities.
Keywords: MYC; medulloblastoma; quantitative proteomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
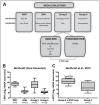
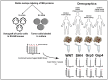
Similar articles
-
Accumulation of genomic aberrations during clinical progression of medulloblastoma.Acta Neuropathol. 2008 Oct;116(4):383-90. doi: 10.1007/s00401-008-0422-y. Epub 2008 Aug 15. Acta Neuropathol. 2008. PMID: 18704466
-
Combined MYC and P53 defects emerge at medulloblastoma relapse and define rapidly progressive, therapeutically targetable disease.Cancer Cell. 2015 Jan 12;27(1):72-84. doi: 10.1016/j.ccell.2014.11.002. Epub 2014 Dec 18. Cancer Cell. 2015. PMID: 25533335 Free PMC article.
-
Somatic cell transfer of c-Myc and Bcl-2 induces large-cell anaplastic medulloblastomas in mice.J Neurooncol. 2016 Feb;126(3):415-24. doi: 10.1007/s11060-015-1985-9. Epub 2015 Oct 30. J Neurooncol. 2016. PMID: 26518543 Free PMC article.
-
Pediatric medulloblastoma in the molecular era: what are the surgical implications?Cancer Metastasis Rev. 2020 Mar;39(1):235-243. doi: 10.1007/s10555-020-09865-y. Cancer Metastasis Rev. 2020. PMID: 32095940 Review.
-
Role of MYC in Medulloblastoma.Cold Spring Harb Perspect Med. 2013 Nov 1;3(11):a014308. doi: 10.1101/cshperspect.a014308. Cold Spring Harb Perspect Med. 2013. PMID: 24186490 Free PMC article. Review.
Cited by
-
HarmonizR enables data harmonization across independent proteomic datasets with appropriate handling of missing values.Nat Commun. 2022 Jun 20;13(1):3523. doi: 10.1038/s41467-022-31007-x. Nat Commun. 2022. PMID: 35725563 Free PMC article.
-
Improved therapy for medulloblastoma: targeting hedgehog and PI3K-mTOR signaling pathways in combination with chemotherapy.Oncotarget. 2018 Mar 30;9(24):16619-16633. doi: 10.18632/oncotarget.24618. eCollection 2018 Mar 30. Oncotarget. 2018. PMID: 29682173 Free PMC article.
-
Identification of key genes and pathways in meningioma by bioinformatics analysis.Oncol Lett. 2018 Jun;15(6):8245-8252. doi: 10.3892/ol.2018.8376. Epub 2018 Mar 29. Oncol Lett. 2018. PMID: 29805558 Free PMC article.
-
Role of MYC-miR-29-B7-H3 in Medulloblastoma Growth and Angiogenesis.J Clin Med. 2019 Aug 2;8(8):1158. doi: 10.3390/jcm8081158. J Clin Med. 2019. PMID: 31382461 Free PMC article.
References
-
- Cho Y.J., Tsherniak A., Tamayo P., Santagata S., Ligon A., Greulich H., Berhoukim R., Amani V., Goumnerova L., Eberhart C.G., et al. Integrative genomic analysis of medulloblastoma identifies a molecular subgroup that drives poor clinical outcome. J. Clin. Oncol. 2011;29:1424–1430. doi: 10.1200/JCO.2010.28.5148. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

