Analysis of the brain mural cell transcriptome
- PMID: 27725773
- PMCID: PMC5057134
- DOI: 10.1038/srep35108
Analysis of the brain mural cell transcriptome
Abstract
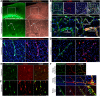
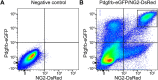
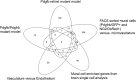
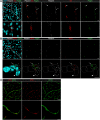
Pericytes, the mural cells of blood microvessels, regulate microvascular development and function and have been implicated in many brain diseases. However, due to a paucity of defining markers, pericyte identification and functional characterization remain ambiguous and data interpretation problematic. In mice carrying two transgenic reporters, Pdgfrb-eGFP and NG2-DsRed, we found that double-positive cells were vascular mural cells, while the single reporters marked additional, but non-overlapping, neuroglial cells. Double-positive cells were isolated by fluorescence-activated cell sorting (FACS) and analyzed by RNA sequencing. To reveal defining patterns of mural cell transcripts, we compared the RNA sequencing data with data from four previously published studies. The meta-analysis provided a conservative catalogue of 260 brain mural cell-enriched gene transcripts. We validated pericyte-specific expression of two novel markers, vitronectin (Vtn) and interferon-induced transmembrane protein 1 (Ifitm1), using fluorescent in situ hybridization and immunohistochemistry. We further analyzed signaling pathways and interaction networks of the pericyte-enriched genes in silico. This work provides novel insight into the molecular composition of brain mural cells. The reported gene catalogue facilitates identification of brain pericytes by providing numerous new candidate marker genes and is a rich source for new hypotheses for future studies of brain mural cell physiology and pathophysiology.
Figures
Similar articles
-
Visualization of vascular mural cells in developing brain using genetically labeled transgenic reporter mice.J Cereb Blood Flow Metab. 2018 Mar;38(3):456-468. doi: 10.1177/0271678X17697720. Epub 2017 Mar 9. J Cereb Blood Flow Metab. 2018. PMID: 28276839 Free PMC article.
-
The complex mural cell: pericyte function in health and disease.Int J Cardiol. 2015;190:75-89. doi: 10.1016/j.ijcard.2015.03.258. Epub 2015 Mar 20. Int J Cardiol. 2015. PMID: 25918055 Review.
-
Microarray analysis of blood microvessels from PDGF-B and PDGF-Rbeta mutant mice identifies novel markers for brain pericytes.FASEB J. 2006 Aug;20(10):1703-5. doi: 10.1096/fj.05-4944fje. Epub 2006 Jun 28. FASEB J. 2006. PMID: 16807374
-
Validation of Specific and Reliable Genetic Tools to Identify, Label, and Target Cardiac Pericytes in Mice.J Am Heart Assoc. 2022 Jan 4;11(1):e023171. doi: 10.1161/JAHA.121.023171. Epub 2021 Dec 22. J Am Heart Assoc. 2022. PMID: 34935413 Free PMC article.
-
Pericytes in Microvessels: From "Mural" Function to Brain and Retina Regeneration.Int J Mol Sci. 2019 Dec 17;20(24):6351. doi: 10.3390/ijms20246351. Int J Mol Sci. 2019. PMID: 31861092 Free PMC article. Review.
Cited by
-
Absence of Cold-Inducible RNA-Binding Protein (CIRP) Promotes Angiogenesis and Regeneration of Ischemic Tissue by Inducing M2-Like Macrophage Polarization.Biomedicines. 2021 Apr 7;9(4):395. doi: 10.3390/biomedicines9040395. Biomedicines. 2021. PMID: 33916904 Free PMC article.
-
Nerve-Glial antigen 2: unmasking the enigmatic cellular identity in the central nervous system.Front Immunol. 2024 Jul 29;15:1393842. doi: 10.3389/fimmu.2024.1393842. eCollection 2024. Front Immunol. 2024. PMID: 39136008 Free PMC article. Review.
-
Transient loss of venous integrity during developmental vascular remodeling leads to red blood cell extravasation and clearance by lymphatic vessels.Development. 2018 Feb 8;145(3):dev156745. doi: 10.1242/dev.156745. Development. 2018. PMID: 29361560 Free PMC article.
-
Transplantation of hPSC-derived pericyte-like cells promotes functional recovery in ischemic stroke mice.Nat Commun. 2020 Oct 15;11(1):5196. doi: 10.1038/s41467-020-19042-y. Nat Commun. 2020. PMID: 33060592 Free PMC article.
-
Whole-brain tissue mapping toolkit using large-scale highly multiplexed immunofluorescence imaging and deep neural networks.Nat Commun. 2021 Mar 10;12(1):1550. doi: 10.1038/s41467-021-21735-x. Nat Commun. 2021. PMID: 33692351 Free PMC article.
References
-
- Sims D. E. The pericyte–a review. Tissue & cell 18, 153–174 (1986). - PubMed
-
- Lindahl P., Johansson B. R., Leveen P. & Betsholtz C. Pericyte loss and microaneurysm formation in PDGF-B-deficient mice. Science 277, 242–245 (1997). - PubMed
-
- Hellstrom M., Kalen M., Lindahl P., Abramsson A. & Betsholtz C. Role of PDGF-B and PDGFR-beta in recruitment of vascular smooth muscle cells and pericytes during embryonic blood vessel formation in the mouse. Development 126, 3047–3055 (1999). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

