Live-cell single-molecule tracking reveals co-recognition of H3K27me3 and DNA targets polycomb Cbx7-PRC1 to chromatin
- PMID: 27723458
- PMCID: PMC5056789
- DOI: 10.7554/eLife.17667
Live-cell single-molecule tracking reveals co-recognition of H3K27me3 and DNA targets polycomb Cbx7-PRC1 to chromatin
Abstract
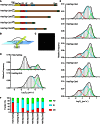
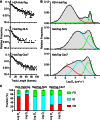
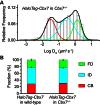
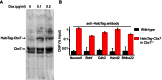
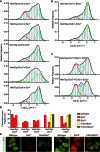
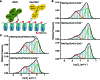
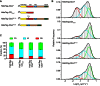
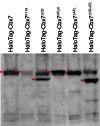
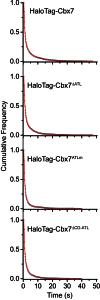
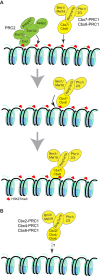
The Polycomb PRC1 plays essential roles in development and disease pathogenesis. Targeting of PRC1 to chromatin is thought to be mediated by the Cbx family proteins (Cbx2/4/6/7/8) binding to histone H3 with a K27me3 modification (H3K27me3). Despite this prevailing view, the molecular mechanisms of targeting remain poorly understood. Here, by combining live-cell single-molecule tracking (SMT) and genetic engineering, we reveal that H3K27me3 contributes significantly to the targeting of Cbx7 and Cbx8 to chromatin, but less to Cbx2, Cbx4, and Cbx6. Genetic disruption of the complex formation of PRC1 facilitates the targeting of Cbx7 to chromatin. Biochemical analyses uncover that the CD and AT-hook-like (ATL) motif of Cbx7 constitute a functional DNA-binding unit. Live-cell SMT of Cbx7 mutants demonstrates that Cbx7 is targeted to chromatin by co-recognizing of H3K27me3 and DNA. Our data suggest a novel hierarchical cooperation mechanism by which histone modifications and DNA coordinate to target chromatin regulatory complexes.
Keywords: Epigenetics; Polycomb; biophysics; chromatin; chromosomes; combinatorial recognition; genes; live-cell imaging; none; single-molecule tracking; structural biology.
Conflict of interest statement
JBG: Filed patent application on the Janelia Fluor (JF) dyes (PCT/US2015/023953). LDL: Filed patent application on the Janelia Fluor (JF) dyes (PCT/US2015/023953). The other authors declare that no competing interests exist.
Figures

Similar articles
-
Nuclear condensates of the Polycomb protein chromobox 2 (CBX2) assemble through phase separation.J Biol Chem. 2019 Feb 1;294(5):1451-1463. doi: 10.1074/jbc.RA118.006620. Epub 2018 Dec 4. J Biol Chem. 2019. PMID: 30514760 Free PMC article.
-
Engagement of DNA and H3K27me3 by the CBX8 chromodomain drives chromatin association.Nucleic Acids Res. 2019 Mar 18;47(5):2289-2305. doi: 10.1093/nar/gky1290. Nucleic Acids Res. 2019. PMID: 30597065 Free PMC article.
-
Optimization of Ligands Using Focused DNA-Encoded Libraries To Develop a Selective, Cell-Permeable CBX8 Chromodomain Inhibitor.ACS Chem Biol. 2020 Jan 17;15(1):112-131. doi: 10.1021/acschembio.9b00654. Epub 2019 Dec 12. ACS Chem Biol. 2020. PMID: 31755685 Free PMC article.
-
Biological functions of chromobox (CBX) proteins in stem cell self-renewal, lineage-commitment, cancer and development.Bone. 2021 Feb;143:115659. doi: 10.1016/j.bone.2020.115659. Epub 2020 Sep 24. Bone. 2021. PMID: 32979540 Review.
-
Beyond EZH2: is the polycomb protein CBX2 an emerging target for anti-cancer therapy?Expert Opin Ther Targets. 2019 Jul;23(7):565-578. doi: 10.1080/14728222.2019.1627329. Epub 2019 Jun 10. Expert Opin Ther Targets. 2019. PMID: 31177918 Review.
Cited by
-
Real-time single-molecule imaging of transcriptional regulatory networks in living cells.Nat Rev Genet. 2024 Apr;25(4):272-285. doi: 10.1038/s41576-023-00684-9. Epub 2024 Jan 9. Nat Rev Genet. 2024. PMID: 38195868 Review.
-
TagBiFC technique allows long-term single-molecule tracking of protein-protein interactions in living cells.Commun Biol. 2021 Mar 19;4(1):378. doi: 10.1038/s42003-021-01896-7. Commun Biol. 2021. PMID: 33742089 Free PMC article.
-
Discovery and Characterization of a Cellular Potent Positive Allosteric Modulator of the Polycomb Repressive Complex 1 Chromodomain, CBX7.Cell Chem Biol. 2019 Oct 17;26(10):1365-1379.e22. doi: 10.1016/j.chembiol.2019.07.013. Epub 2019 Aug 15. Cell Chem Biol. 2019. PMID: 31422906 Free PMC article.
-
Protein motion in the nucleus: from anomalous diffusion to weak interactions.Biochem Soc Trans. 2018 Aug 20;46(4):945-956. doi: 10.1042/BST20170310. Epub 2018 Jul 31. Biochem Soc Trans. 2018. PMID: 30065106 Free PMC article. Review.
-
Epigenetic regulatory layers in the 3D nucleus.Mol Cell. 2024 Feb 1;84(3):415-428. doi: 10.1016/j.molcel.2023.12.032. Epub 2024 Jan 18. Mol Cell. 2024. PMID: 38242127 Review.
References
-
- Akasaka T, Kanno M, Balling R, Mieza MA, Taniguchi M, Koseki H. A role for mel-18, a Polycomb group-related vertebrate gene, during theanteroposterior specification of the axial skeleton. Development. 1996;122:1513–1522. - PubMed
-
- Blackledge NP, Farcas AM, Kondo T, King HW, McGouran JF, Hanssen LL, Ito S, Cooper S, Kondo K, Koseki Y, Ishikura T, Long HK, Sheahan TW, Brockdorff N, Kessler BM, Koseki H, Klose RJ. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell. 2014;157:1445–1459. doi: 10.1016/j.cell.2014.05.004. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

