Rapid change of fecal microbiome and disappearance of Clostridium difficile in a colonized infant after transition from breast milk to cow milk
- PMID: 27717398
- PMCID: PMC5055705
- DOI: 10.1186/s40168-016-0198-6
Rapid change of fecal microbiome and disappearance of Clostridium difficile in a colonized infant after transition from breast milk to cow milk
Abstract
Background: Clostridium difficile is the most common known cause of antibiotic-associated diarrhea. Upon the disturbance of gut microbiota by antibiotics, C. difficile establishes growth and releases toxins A and B, which cause tissue damage in the host. The symptoms of C. difficile infection disease range from mild diarrhea to pseudomembranous colitis and toxic megacolon. Interestingly, 10-50 % of infants are asymptomatic carriers of C. difficile. This longitudinal study of the C. difficile colonization in an infant revealed the dynamics of C. difficile presence in gut microbiota.
Methods: Fifty fecal samples, collected weekly between 5.5 and 17 months of age from a female infant who was an asymptomatic carrier of C. difficile, were analyzed by 16S rRNA gene sequencing.
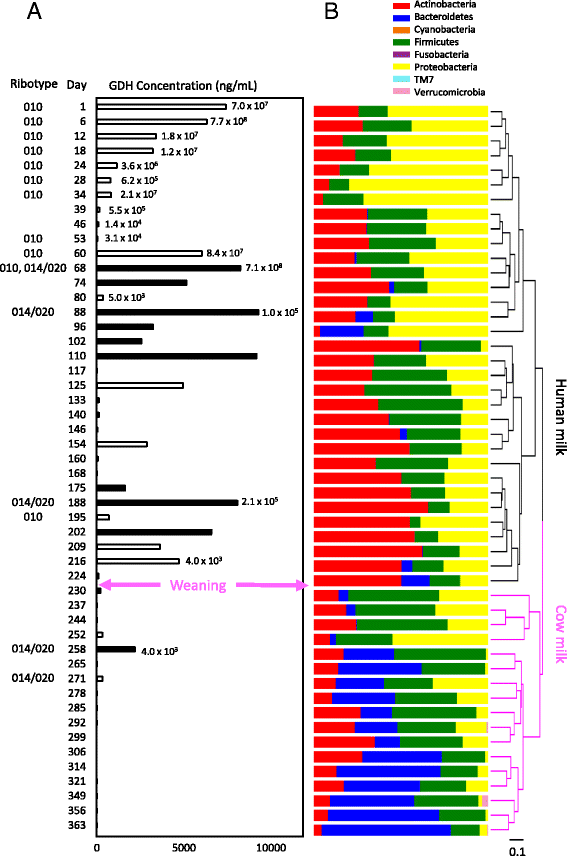
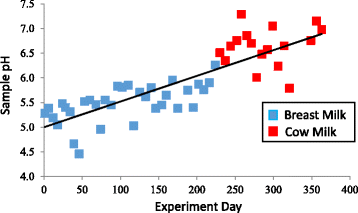
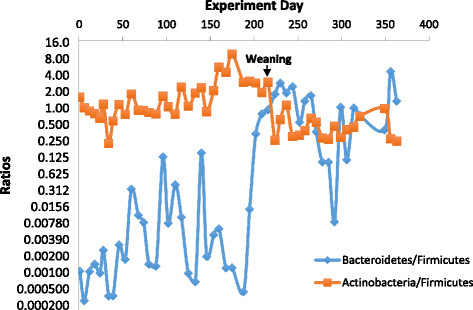
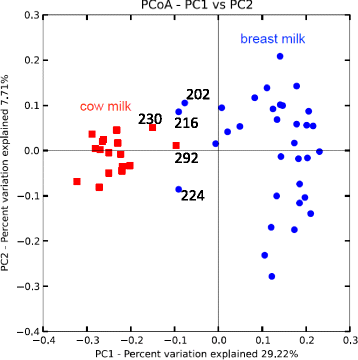
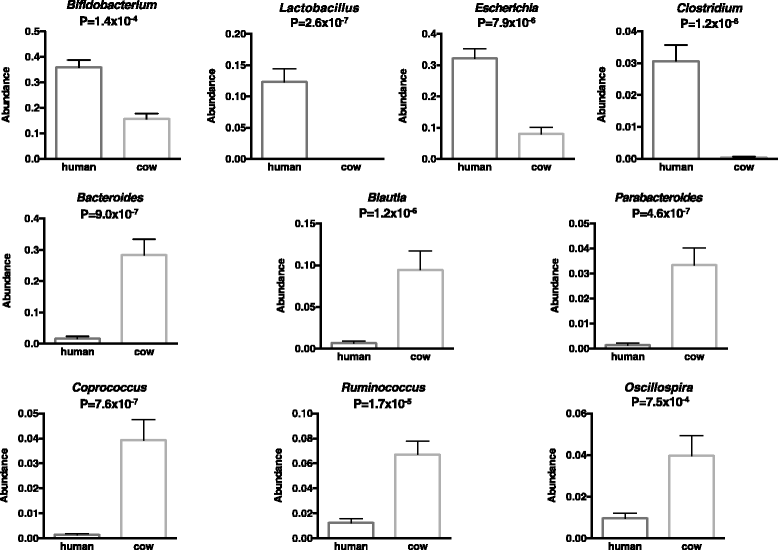
Results: Colonization switching between toxigenic and non-toxigenic C. difficile strains as well as more than 100,000-fold fluctuations of C. difficile counts were observed. C. difficile toxins were detected during the testing period in some infant stool samples, but the infant never had diarrhea. Although fecal microbiota was stable during breast feeding, a dramatic and permanent change of microbiota composition was observed within 5 days of the transition from human milk to cow milk. A rapid decline and eventual disappearance of C. difficile coincided with weaning at 12.5 months. An increase in the relative abundance of Bacteroides spp., Blautia spp., Parabacteroides spp., Coprococcus spp., Ruminococcus spp., and Oscillospira spp. and a decrease of Bifidobacterium spp., Lactobacillus spp., Escherichia spp., and Clostridium spp. were observed during weaning. The change in microbiome composition was accompanied by a gradual increase of fecal pH from 5.5 to 7.
Conclusions: The bacterial groups that are less abundant in early infancy, and that increase in relative abundance after weaning, likely are responsible for the expulsion of C. difficile.
Keywords: C. difficile; Human milk; Infant gut microbiome.
Figures
Similar articles
-
Insight into alteration of gut microbiota in Clostridium difficile infection and asymptomatic C. difficile colonization.Anaerobe. 2015 Aug;34:1-7. doi: 10.1016/j.anaerobe.2015.03.008. Epub 2015 Mar 26. Anaerobe. 2015. PMID: 25817005
-
Fecal Microbiome Among Nursing Home Residents with Advanced Dementia and Clostridium difficile.Dig Dis Sci. 2018 Jun;63(6):1525-1531. doi: 10.1007/s10620-018-5030-7. Epub 2018 Mar 28. Dig Dis Sci. 2018. PMID: 29594967 Free PMC article.
-
Gut dysbiosis following C-section instigates higher colonisation of toxigenic Clostridium perfringens in infants.Benef Microbes. 2017 May 30;8(3):353-365. doi: 10.3920/BM2016.0216. Epub 2017 May 15. Benef Microbes. 2017. PMID: 28504574
-
Theodore E. Woodward Award. How bacterial enterotoxins work: insights from in vivo studies.Trans Am Clin Climatol Assoc. 2002;113:167-80; discussion 180-1. Trans Am Clin Climatol Assoc. 2002. PMID: 12053708 Free PMC article. Review.
-
Asymptomatic Clostridium difficile colonization: epidemiology and clinical implications.BMC Infect Dis. 2015 Nov 14;15:516. doi: 10.1186/s12879-015-1258-4. BMC Infect Dis. 2015. PMID: 26573915 Free PMC article. Review.
Cited by
-
The Comparative Analysis of Genomic Diversity and Genes Involved in Carbohydrate Metabolism of Eighty-Eight Bifidobacterium pseudocatenulatum Isolates from Different Niches of China.Nutrients. 2022 Jun 4;14(11):2347. doi: 10.3390/nu14112347. Nutrients. 2022. PMID: 35684146 Free PMC article.
-
The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota.Microbiol Mol Biol Rev. 2017 Nov 8;81(4):e00036-17. doi: 10.1128/MMBR.00036-17. Print 2017 Dec. Microbiol Mol Biol Rev. 2017. PMID: 29118049 Free PMC article. Review.
-
Low diversity gut microbiota dysbiosis: drivers, functional implications and recovery.Curr Opin Microbiol. 2018 Aug;44:34-40. doi: 10.1016/j.mib.2018.07.003. Epub 2018 Jul 20. Curr Opin Microbiol. 2018. PMID: 30036705 Free PMC article. Review.
-
Comparing Gut Microbiome in Mothers' Own Breast Milk- and Formula-Fed Moderate-Late Preterm Infants.Front Microbiol. 2020 May 26;11:891. doi: 10.3389/fmicb.2020.00891. eCollection 2020. Front Microbiol. 2020. PMID: 32528425 Free PMC article.
-
Neurodevelopmental Disorders Associated with Gut Microbiome Dysbiosis in Children.Children (Basel). 2024 Jun 28;11(7):796. doi: 10.3390/children11070796. Children (Basel). 2024. PMID: 39062245 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

