Evasion of early innate immune response by 2'-O-methylation of dengue genomic RNA
- PMID: 27716465
- PMCID: PMC7172056
- DOI: 10.1016/j.virol.2016.09.022
Evasion of early innate immune response by 2'-O-methylation of dengue genomic RNA
Abstract
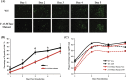
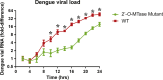
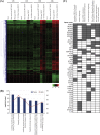
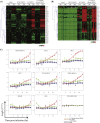
Dengue virus (DENV) is the most prevalent mosquito-borne virus pathogen in humans. There is currently no antiviral therapeutic or widely available vaccine against dengue infection. The DENV RNA genome is methylated on its 5' cap by its NS5 protein. DENV bearing a single E216A point mutation in NS5 loses 2'-O-methylation of its genome. While this mutant DENV is highly attenuated and immunogenic, the mechanism of this attenuation has not been elucidated. In this study, we find that replication of this mutant DENV is attenuated very early during infection. This early attenuation is not dependent on a functional type I interferon response and coincides with early activation of the innate immune response. Taken together, our data suggest that 2'-O-methylation of DENV genomic RNA is important for evasion of the host immune response during the very early stages of infection as the virus seeks to establish infection.
Keywords: Dengue virus; Innate immune response; Methyltransferase; NS5.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures
Similar articles
-
Molecular basis for specific viral RNA recognition and 2'-O-ribose methylation by the dengue virus nonstructural protein 5 (NS5).Proc Natl Acad Sci U S A. 2015 Dec 1;112(48):14834-9. doi: 10.1073/pnas.1514978112. Epub 2015 Nov 17. Proc Natl Acad Sci U S A. 2015. PMID: 26578813 Free PMC article.
-
Live Cell Analysis and Mathematical Modeling Identify Determinants of Attenuation of Dengue Virus 2'-O-Methylation Mutant.PLoS Pathog. 2015 Dec 31;11(12):e1005345. doi: 10.1371/journal.ppat.1005345. eCollection 2015 Dec. PLoS Pathog. 2015. PMID: 26720415 Free PMC article.
-
How Dengue Virus Circumvents Innate Immunity.Front Immunol. 2018 Dec 4;9:2860. doi: 10.3389/fimmu.2018.02860. eCollection 2018. Front Immunol. 2018. PMID: 30564245 Free PMC article. Review.
-
Innate Antiviral Immunity against Dengue Virus.Crit Rev Immunol. 2015;35(3):253-60. doi: 10.1615/critrevimmunol.2015014251. Crit Rev Immunol. 2015. PMID: 26559229 Review.
-
Innate immunity to dengue virus infection and subversion of antiviral responses.J Mol Biol. 2014 Mar 20;426(6):1148-60. doi: 10.1016/j.jmb.2013.11.023. Epub 2013 Dec 3. J Mol Biol. 2014. PMID: 24316047 Free PMC article. Review.
Cited by
-
Biochemistry and Molecular Biology of Flaviviruses.Chem Rev. 2018 Apr 25;118(8):4448-4482. doi: 10.1021/acs.chemrev.7b00719. Epub 2018 Apr 13. Chem Rev. 2018. PMID: 29652486 Free PMC article. Review.
-
The hidden RNA code: implications of the RNA epitranscriptome in the context of viral infections.Front Genet. 2023 Aug 1;14:1245683. doi: 10.3389/fgene.2023.1245683. eCollection 2023. Front Genet. 2023. PMID: 37614818 Free PMC article. Review.
-
The Emerging Role of RNA Modifications in the Regulation of Antiviral Innate Immunity.Front Microbiol. 2022 Feb 3;13:845625. doi: 10.3389/fmicb.2022.845625. eCollection 2022. Front Microbiol. 2022. PMID: 35185855 Free PMC article. Review.
-
New Targets for Zika Virus Determined by Human-Viral Interactomic: A Bioinformatics Approach.Biomed Res Int. 2017;2017:1734151. doi: 10.1155/2017/1734151. Epub 2017 Dec 12. Biomed Res Int. 2017. PMID: 29379794 Free PMC article.
-
A conserved arginine in NS5 binds genomic 3' stem-loop RNA for primer-independent initiation of flavivirus RNA replication.RNA. 2022 Feb;28(2):177-193. doi: 10.1261/rna.078949.121. Epub 2021 Nov 10. RNA. 2022. PMID: 34759006 Free PMC article.
References
-
- Ackermann M., Padmanabhan R. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J. Biol. Chem. 2001;276(43):39926–39937. - PubMed
-
- Bhatt S., Gething P.W., Brady O.J., Messina J.P., Farlow A.W., Moyes C.L., Drake J.M., Brownstein J.S., Hoen A.G., Sankoh O., Myers M.F., George D.B., Jaenisch T., Wint G.R., Simmons C.P., Scott T.W., Farrar J.J., Hay S.I. The global distribution and burden of dengue. Nature. 2013;496(7446):504–507. - PMC - PubMed
-
- Daffis S., Szretter K.J., Schriewer J., Li J., Youn S., Errett J., Lin T.Y., Schneller S., Zust R., Dong H., Thiel V., Sen G.C., Fensterl V., Klimstra W.B., Pierson T.C., Buller R.M., Gale M., Jr., Shi P.Y., Diamond M.S. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 2010;468(7322):452–456. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

