Characterization of virus-derived small interfering RNAs in Apple stem grooving virus-infected in vitro-cultured Pyrus pyrifolia shoot tips in response to high temperature treatment
- PMID: 27716257
- PMCID: PMC5053029
- DOI: 10.1186/s12985-016-0625-0
Characterization of virus-derived small interfering RNAs in Apple stem grooving virus-infected in vitro-cultured Pyrus pyrifolia shoot tips in response to high temperature treatment
Abstract
Background: Heat treatment (known as thermotherapy) together with in vitro culture of shoot meristem tips is a commonly used technology to obtain virus-free germplasm for the effective control of virus diseases in fruit trees. RNA silencing as an antiviral defense mechanism has been implicated in this process. To understand if high temperature-mediated acceleration of the host antiviral gene silencing system in the meristem tip facilitates virus-derived small interfering RNAs (vsiRNA) accumulation to reduce the viral RNA titer in the fruit tree meristem tip cells, we used the Apple stem grooving virus (ASGV)-Pyrus pyrifolia pathosystem to explore the possible roles of vsiRNA in thermotherapy.
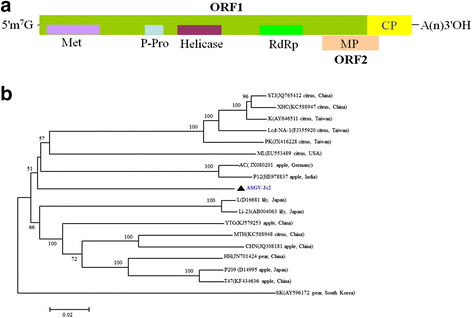
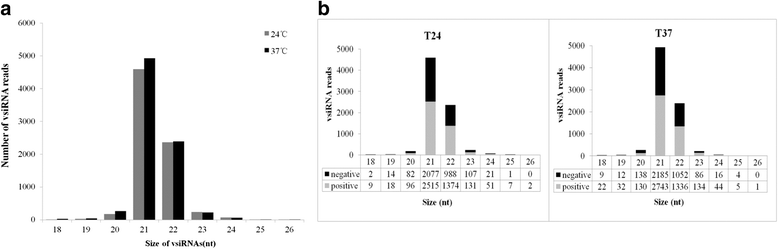
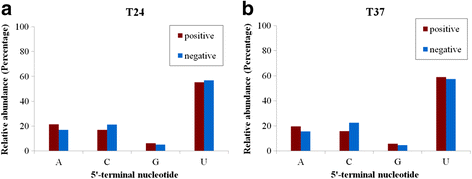
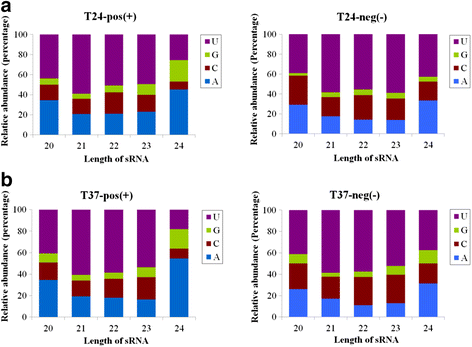
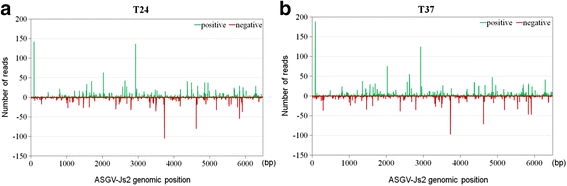
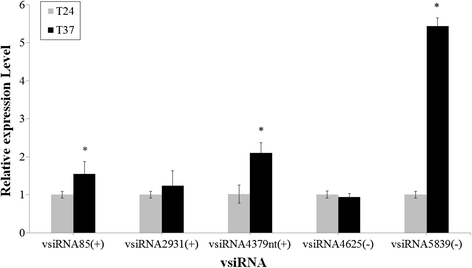
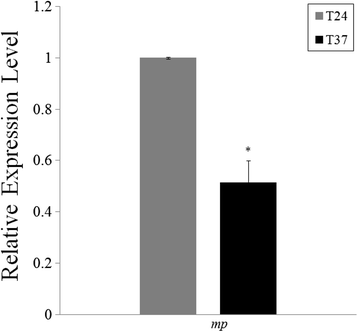
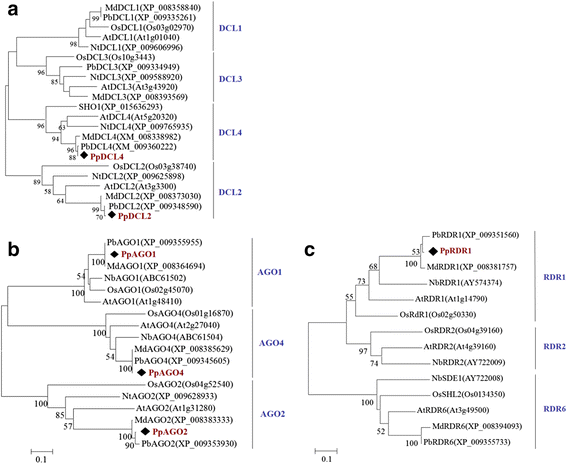
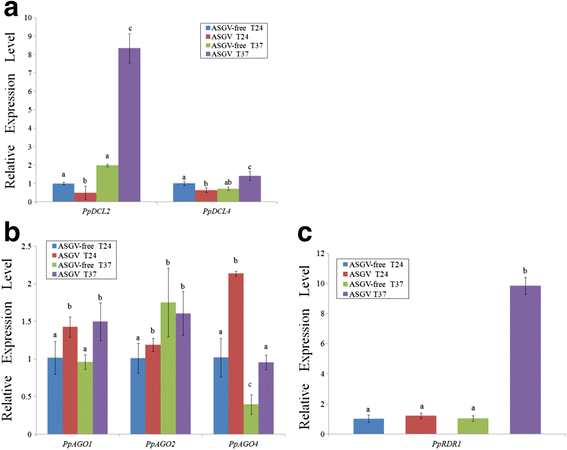
Results: At first we determined the full-length genome sequence of the ASGV-Js2 isolate and then profiled vsiRNAs in the meristem tip of in vitro-grown pear (cv. 'Jinshui no. 2') shoots infected by ASGV-Js2 and cultured at 24 and 37 °C. A total of 7,495 and 7,949 small RNA reads were obtained from the tips of pear shoots cultured at 24 and 37 °C, respectively. Mapping of the vsiRNAs to the ASGV-Js2 genome revealed that they were unevenly distributed along the ASGV-Js2 genome, and that 21- and 22-nt vsiRNAs preferentially accumulated at both temperatures. The 5'-terminal nucleotides of ASGV-specific siRNAs in the tips cultured under different temperatures had a similar distribution pattern, and the nucleotide U was the most frequent. RT-qPCR analyses suggested that viral genome accumulation was drastically compromised at 37 °C compared to 24 °C, which was accompanied with the elevated levels of vsiRNAs at 37 °C. As plant Dicer-like proteins (DCLs), Argonaute proteins (AGOs), and RNA-dependent RNA polymerases (RDRs) are implicated in vsiRNA biogenesis, we also cloned the partial sequences of PpDCL2,4, PpAGO1,2,4 and PpRDR1 genes, and found their expression levels were up-regulated in the ASGV-infected pear shoots at 37 °C.
Conclusions: Collectively, these results showed that upon high temperature treatment, the ASGV-infected meristem shoot tips up-regulated the expression of key genes in the RNA silencing pathway, induced the biogenesis of vsiRNAs and inhibited viral RNA accumulation. This study represents the first report on the characterization of the vsiRNA population in pear plants infected by ASGV-Js2, in response to high temperature treatment.
Keywords: Apple stem grooving virus; Argonaute (AGO); Dicer-like (DCL); Gene silencing; High temperature; Pyrus pyrifolia; RNA dependent RNA polymerase (RDRs); RT-qPCR; Virus-derived small interfering RNA (vsiRNA).
Figures
Similar articles
-
Identification and characterization of microRNAs from in vitro-grown pear shoots infected with Apple stem grooving virus in response to high temperature using small RNA sequencing.BMC Genomics. 2015 Nov 16;16:945. doi: 10.1186/s12864-015-2126-8. BMC Genomics. 2015. PMID: 26573813 Free PMC article.
-
Combining Thermotherapy with Cryotherapy for Efficient Eradication of Apple stem grooving virus from Infected In-vitro-cultured Apple Shoots.Plant Dis. 2018 Aug;102(8):1574-1580. doi: 10.1094/PDIS-11-17-1753-RE. Epub 2018 Jun 11. Plant Dis. 2018. PMID: 30673422
-
Characteristics of siRNAs derived from Southern rice black-streaked dwarf virus in infected rice and their potential role in host gene regulation.Virol J. 2017 Feb 10;14(1):27. doi: 10.1186/s12985-017-0699-3. Virol J. 2017. PMID: 28183327 Free PMC article.
-
Biogenesis, Function, and Applications of Virus-Derived Small RNAs in Plants.Front Microbiol. 2015 Nov 9;6:1237. doi: 10.3389/fmicb.2015.01237. eCollection 2015. Front Microbiol. 2015. PMID: 26617580 Free PMC article. Review.
-
Recent progress on gene silencing/suppression by virus-derived small interfering RNAs in rice viruses especially Rice grassy stunt virus.Microb Pathog. 2018 Dec;125:210-218. doi: 10.1016/j.micpath.2018.09.021. Epub 2018 Sep 19. Microb Pathog. 2018. PMID: 30243549 Review.
Cited by
-
Profile of siRNAs derived from green fluorescent protein (GFP)-tagged Papaya leaf distortion mosaic virus in infected papaya plants.Virus Genes. 2018 Dec;54(6):833-839. doi: 10.1007/s11262-018-1601-0. Epub 2018 Sep 14. Virus Genes. 2018. PMID: 30218292
-
In vitro thermotherapy-based methods for plant virus eradication.Plant Methods. 2018 Oct 6;14:87. doi: 10.1186/s13007-018-0355-y. eCollection 2018. Plant Methods. 2018. PMID: 30323856 Free PMC article. Review.
-
Elevated Carbon Dioxide Levels Decreases Cucumber Mosaic Virus Accumulation in Correlation with Greater Accumulation of rgs-CaM, an Inhibitor of a Viral Suppressor of RNAi.Plants (Basel). 2020 Dec 29;10(1):59. doi: 10.3390/plants10010059. Plants (Basel). 2020. PMID: 33383811 Free PMC article.
-
Elimination of Eight Viruses and Two Viroids from Preclonal Candidates of Six Grapevine Varieties (Vitis vinifera L.) through In Vivo Thermotherapy and In Vitro Meristem Tip Micrografting.Plants (Basel). 2022 Apr 13;11(8):1064. doi: 10.3390/plants11081064. Plants (Basel). 2022. PMID: 35448791 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

