Metal-Induced Stabilization and Activation of Plasmid Replication Initiator RepB
- PMID: 27709114
- PMCID: PMC5030251
- DOI: 10.3389/fmolb.2016.00056
Metal-Induced Stabilization and Activation of Plasmid Replication Initiator RepB
Abstract
Initiation of plasmid rolling circle replication (RCR) is catalyzed by a plasmid-encoded Rep protein that performs a Tyr- and metal-dependent site-specific cleavage of one DNA strand within the double-strand origin (dso) of replication. The crystal structure of RepB, the initiator protein of the streptococcal plasmid pMV158, constitutes the first example of a Rep protein structure from RCR plasmids. It forms a toroidal homohexameric ring where each RepB protomer consists of two domains: the C-terminal domain involved in oligomerization and the N-terminal domain containing the DNA-binding and endonuclease activities. Binding of Mn2+ to the active site is essential for the catalytic activity of RepB. In this work, we have studied the effects of metal binding on the structure and thermostability of full-length hexameric RepB and each of its separate domains by using different biophysical approaches. The analysis of the temperature-induced changes in RepB shows that the first thermal transition, which occurs at a range of temperatures physiologically relevant for the pMV158 pneumococcal host, represents an irreversible conformational change that affects the secondary and tertiary structure of the protein, which becomes prone to self-associate. This transition, which is also shown to result in loss of DNA binding capacity and catalytic activity of RepB, is confined to its N-terminal domain. Mn2+ protects the protein from undergoing this detrimental conformational change and the observed protection correlates well with the high-affinity binding of the cation to the active site, as substituting one of the metal-ligands at this site impairs both the protein affinity for Mn2+and the Mn2+-driven thermostabilization effect. The level of catalytic activity of the protein, especially in the case of full-length RepB, cannot be explained based only on the high-affinity binding of Mn2+ at the active site and suggests the existence of additional, lower-affinity metal binding site(s), missing in the separate catalytic domain, that must also be saturated for maximal activity. The molecular bases of the thermostabilizing effect of Mn2+ on the N-terminal domain of the protein as well as the potential location of additional metal binding sites in the entire RepB are discussed.
Keywords: HUH endonucleases; Mn2+ affinity; RepB thermostability; metal-dependent catalytic activity; plasmid-encoded Rep proteins.
Figures
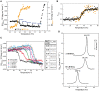
 ) between 10 and 60°C. The solid line shows the fit of Equation (1) to FDapp values. (C) Temperature transition curves of RepB6 (12 μM) in the presence of increasing concentrations of MnCl2 (indicated inside the graph) measured by CD at 218 nm. The table shows the apparent half-transition temperatures of RepB6 derived from fit of Equation (1) to the figure experimental curves (solid lines). (D) DSC profile of the first thermal transition of RepB6 (30 μM) monitored in the absence and in the presence of 130 μM or 2 mM Mn2+. The position of the maximum of the heat capacity function (Tm) is indicated.
) between 10 and 60°C. The solid line shows the fit of Equation (1) to FDapp values. (C) Temperature transition curves of RepB6 (12 μM) in the presence of increasing concentrations of MnCl2 (indicated inside the graph) measured by CD at 218 nm. The table shows the apparent half-transition temperatures of RepB6 derived from fit of Equation (1) to the figure experimental curves (solid lines). (D) DSC profile of the first thermal transition of RepB6 (30 μM) monitored in the absence and in the presence of 130 μM or 2 mM Mn2+. The position of the maximum of the heat capacity function (Tm) is indicated.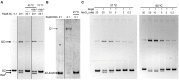

 ), OBD (
), OBD ( ), and OD (
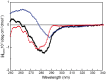
), and OD ( ) separate domains. [Θ] represents the mean residue ellipticity. For comparison, the OBD and OD spectra have been weighted by the fractional contribution of their amino acids to the complete protein sequence.
) separate domains. [Θ] represents the mean residue ellipticity. For comparison, the OBD and OD spectra have been weighted by the fractional contribution of their amino acids to the complete protein sequence.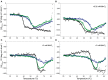
 ), and OBDD42A (
), and OBDD42A ( ) measured by CD at 218 nm in the absence of MnCl2
(A) or in the presence of 0.05 (B), 2 (C), and 5 (D) mM of MnCl2 ([Θ] represents the protein molar ellipticity). The CD thermal profile of RepB6 has been shifted along the ordinate axis to facilitate the comparison with those OBD and OBDD42A. In the case of OBD and OBDD42A, the continuous lines represent an average of the experimental data. Measurements were carried out at 12 μM RepB6 and 19 μM OBD or OBDD42A.
) measured by CD at 218 nm in the absence of MnCl2
(A) or in the presence of 0.05 (B), 2 (C), and 5 (D) mM of MnCl2 ([Θ] represents the protein molar ellipticity). The CD thermal profile of RepB6 has been shifted along the ordinate axis to facilitate the comparison with those OBD and OBDD42A. In the case of OBD and OBDD42A, the continuous lines represent an average of the experimental data. Measurements were carried out at 12 μM RepB6 and 19 μM OBD or OBDD42A.

 ) and OBDD42A (
) and OBDD42A ( ) is indicated on the y-axis.
) is indicated on the y-axis.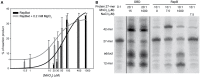
 ) is indicated on the y-axis. (B) Reaction product pattern generated by the nicking and strand-transfer activities of RepB6 and OBD on ssDNA oligos. The assays were performed as those depicted in (A), at the protein:oligo substrate molar ratio and MnCl2 concentration indicated on the top of each lane. As a control, the reaction was also carried out with NaCl instead of manganese salt. The resultant fluorescent oligos were analyzed, visualized and quantified as in Figure 7. To compare the reactions products generated by the activity of OBD and RepB6 the images from different gels acquired and processed under the same conditions have been grouped and indicated by dividing lines.
) is indicated on the y-axis. (B) Reaction product pattern generated by the nicking and strand-transfer activities of RepB6 and OBD on ssDNA oligos. The assays were performed as those depicted in (A), at the protein:oligo substrate molar ratio and MnCl2 concentration indicated on the top of each lane. As a control, the reaction was also carried out with NaCl instead of manganese salt. The resultant fluorescent oligos were analyzed, visualized and quantified as in Figure 7. To compare the reactions products generated by the activity of OBD and RepB6 the images from different gels acquired and processed under the same conditions have been grouped and indicated by dividing lines.Similar articles
-
Acidic pH Decreases the Endonuclease Activity of Initiator RepB and Increases the Stability of the Covalent RepB-DNA Intermediate while Has Only a Limited Effect on the Replication of Plasmid pMV158 in Lactococcus lactis.Front Mol Biosci. 2021 Mar 5;8:634461. doi: 10.3389/fmolb.2021.634461. eCollection 2021. Front Mol Biosci. 2021. PMID: 33889596 Free PMC article.
-
Structures of pMV158 replication initiator RepB with and without DNA reveal a flexible dual-function protein.Nucleic Acids Res. 2023 Feb 22;51(3):1458-1472. doi: 10.1093/nar/gkac1271. Nucleic Acids Res. 2023. PMID: 36688326 Free PMC article.
-
Plasmid replication initiator RepB forms a hexamer reminiscent of ring helicases and has mobile nuclease domains.EMBO J. 2009 Jun 3;28(11):1666-78. doi: 10.1038/emboj.2009.125. EMBO J. 2009. PMID: 19440202 Free PMC article.
-
Plasmid rolling-circle replication: highlights of two decades of research.Plasmid. 2005 Mar;53(2):126-36. doi: 10.1016/j.plasmid.2004.12.008. Epub 2005 Jan 22. Plasmid. 2005. PMID: 15737400 Review.
-
DNA-protein interactions during the initiation and termination of plasmid pT181 rolling-circle replication.Prog Nucleic Acid Res Mol Biol. 2003;75:113-37. doi: 10.1016/s0079-6603(03)75004-1. Prog Nucleic Acid Res Mol Biol. 2003. PMID: 14604011 Review.
Cited by
-
The Different Faces of Rolling-Circle Replication and Its Multifunctional Initiator Proteins.Front Microbiol. 2017 Nov 30;8:2353. doi: 10.3389/fmicb.2017.02353. eCollection 2017. Front Microbiol. 2017. PMID: 29250047 Free PMC article. Review.
-
From Data Mining of Chitinophaga sp. Genome to Enzyme Discovery of a Hyperthermophilic Metallocarboxypeptidase.Microorganisms. 2021 Feb 14;9(2):393. doi: 10.3390/microorganisms9020393. Microorganisms. 2021. PMID: 33673011 Free PMC article.
-
Acidic pH Decreases the Endonuclease Activity of Initiator RepB and Increases the Stability of the Covalent RepB-DNA Intermediate while Has Only a Limited Effect on the Replication of Plasmid pMV158 in Lactococcus lactis.Front Mol Biosci. 2021 Mar 5;8:634461. doi: 10.3389/fmolb.2021.634461. eCollection 2021. Front Mol Biosci. 2021. PMID: 33889596 Free PMC article.
-
Replication of Staphylococcal Resistance Plasmids.Front Microbiol. 2017 Nov 23;8:2279. doi: 10.3389/fmicb.2017.02279. eCollection 2017. Front Microbiol. 2017. PMID: 29218034 Free PMC article. Review.
References
-
- Boer R., Russi S., Guasch A., Lucas M., Blanco A. G., Pérez-Luque R., et al. . (2006). Unveiling the molecular mechanism of a conjugative relaxase: the structure of TrwC complexed with a 27-mer DNA comprising the recognition hairpin and the cleavage site. J. Mol. Biol. 358, 857. 10.1016/j.jmb.2006.02.018 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

