Local modulation of chemoattractant concentrations by single cells: dissection using a bulk-surface computational model
- PMID: 27708760
- PMCID: PMC4992739
- DOI: 10.1098/rsfs.2016.0036
Local modulation of chemoattractant concentrations by single cells: dissection using a bulk-surface computational model
Abstract
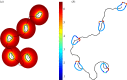
Chemoattractant gradients are usually considered in terms of sources and sinks that are independent of the chemotactic cell. However, recent interest has focused on 'self-generated' gradients, in which cell populations create their own local gradients as they move. Here, we consider the interplay between chemoattractants and single cells. To achieve this, we extend a recently developed computational model to incorporate breakdown of extracellular attractants by membrane-bound enzymes. Model equations are parametrized, using the published estimates from Dictyostelium cells chemotaxing towards cyclic AMP. We find that individual cells can substantially modulate their local attractant field under physiologically appropriate conditions of attractant and enzymes. This means the attractant concentration perceived by receptors can be a small fraction of the ambient concentration. This allows efficient chemotaxis in chemoattractant concentrations that would be saturating without local breakdown. Similar interactions in which cells locally mould a stimulus could function in many types of directed cell motility, including haptotaxis, durotaxis and even electrotaxis.
Keywords: bulk-surface model; cell motility; chemotaxis; self-generated gradients; surface finite-elements.
Figures
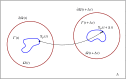
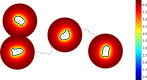
 with far-field boundary
with far-field boundary  . After a time interval of size Δt, the material point located at
. After a time interval of size Δt, the material point located at  on the cell membrane Γ(t) evolves to the new location
on the cell membrane Γ(t) evolves to the new location  .
.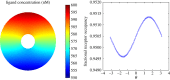
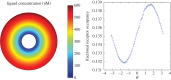
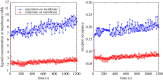
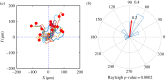
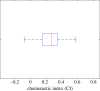
 and the standard error of the mean
and the standard error of the mean  .
.
Similar articles
-
Self-Generated Chemoattractant Gradients: Attractant Depletion Extends the Range and Robustness of Chemotaxis.PLoS Biol. 2016 Mar 16;14(3):e1002404. doi: 10.1371/journal.pbio.1002404. eCollection 2016 Mar. PLoS Biol. 2016. PMID: 26981861 Free PMC article.
-
Self-Generated Gradients Yield Exceptionally Robust Steering Cues.Front Cell Dev Biol. 2020 Mar 5;8:133. doi: 10.3389/fcell.2020.00133. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32195256 Free PMC article.
-
Pseudopodium activation and inhibition signals in chemotaxis by Dictyostelium discoideum amoebae.Semin Cell Biol. 1990 Apr;1(2):87-97. Semin Cell Biol. 1990. PMID: 2102388 Review.
-
Seeing around corners: Cells solve mazes and respond at a distance using attractant breakdown.Science. 2020 Aug 28;369(6507):eaay9792. doi: 10.1126/science.aay9792. Science. 2020. PMID: 32855311
-
Steering yourself by the bootstraps: how cells create their own gradients for chemotaxis.Trends Cell Biol. 2022 Jul;32(7):585-596. doi: 10.1016/j.tcb.2022.02.007. Epub 2022 Mar 26. Trends Cell Biol. 2022. PMID: 35351380 Review.
Cited by
-
Three-component contour dynamics model to simulate and analyze amoeboid cell motility in two dimensions.PLoS One. 2024 Jan 26;19(1):e0297511. doi: 10.1371/journal.pone.0297511. eCollection 2024. PLoS One. 2024. PMID: 38277351 Free PMC article.
-
An Adaptive Moving Mesh Method for Forced Curve Shortening Flow.SIAM J Sci Comput. 2019;41(2):A1170-A1200. doi: 10.1137/18M1211969. SIAM J Sci Comput. 2019. PMID: 31798297 Free PMC article.
-
Chemotactic network responses to live bacteria show independence of phagocytosis from chemoreceptor sensing.Elife. 2017 May 25;6:e24627. doi: 10.7554/eLife.24627. Elife. 2017. PMID: 28541182 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

