The Identification of a Novel Mutant Allele of topoisomerase II in Caenorhabditis elegans Reveals a Unique Role in Chromosome Segregation During Spermatogenesis
- PMID: 27707787
- PMCID: PMC5161275
- DOI: 10.1534/genetics.116.195099
The Identification of a Novel Mutant Allele of topoisomerase II in Caenorhabditis elegans Reveals a Unique Role in Chromosome Segregation During Spermatogenesis
Abstract
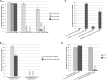
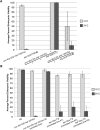
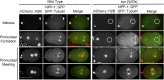
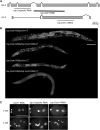
Topoisomerase II alleviates DNA entanglements that are generated during mitotic DNA replication, transcription, and sister chromatid separation. In contrast to mitosis, meiosis has two rounds of chromosome segregation following one round of DNA replication. In meiosis II, sister chromatids segregate from each other, similar to mitosis. Meiosis I, on the other hand, segregates homologs, which requires pairing, synapsis, and recombination. The exact role that topoisomerase II plays during meiosis is unknown. In a screen reexamining Caenorhabditis elegans legacy mutants isolated 30 years ago, we identified a novel allele of the gene encoding topoisomerase II, top-2(it7). In this study, we demonstrate that top-2(it7) males produce dead embryos, even when fertilizing wild-type oocytes. Characterization of early embryonic events indicates that fertilization is successful and sperm components are transmitted to the embryo. However, sperm chromatin is not detected in these fertilized embryos. Examination of top-2(it7) spermatogenic germ lines reveals that the sperm DNA fails to segregate properly during anaphase I of meiosis, resulting in anucleate sperm. top-2(it7) chromosome-segregation defects observed during anaphase I are not due to residual entanglements incurred during meiotic DNA replication and are not dependent on SPO-11-induced double-strand DNA breaks. Finally, we show that TOP-2 associates with chromosomes in meiotic prophase and that chromosome association is disrupted in the germ lines of top-2(it7) mutants.
Keywords: Caenorhabditis elegans; meiosis; spermatogenesis; top-2; topoisomerase II.
Copyright © 2016 by the Genetics Society of America.
Figures

Similar articles
-
Mutations in tyrosyl-DNA phosphodiesterase 2 suppress top-2 induced chromosome segregation defects during Caenorhabditis elegans spermatogenesis.J Biol Chem. 2024 Jul;300(7):107446. doi: 10.1016/j.jbc.2024.107446. Epub 2024 Jun 4. J Biol Chem. 2024. PMID: 38844130 Free PMC article.
-
TOP-2 is differentially required for the proper maintenance of the cohesin subunit REC-8 on meiotic chromosomes in Caenorhabditis elegans spermatogenesis and oogenesis.Genetics. 2022 Sep 30;222(2):iyac120. doi: 10.1093/genetics/iyac120. Genetics. 2022. PMID: 35951744 Free PMC article.
-
New Role for an Old Protein: An Educational Primer for Use with "The Identification of a Novel Mutant Allele of topoisomerase II in Caenorhabditis elegans Reveals a Unique Role in Chromosome Segregation During Spermatogenesis".Genetics. 2018 Jan;208(1):79-88. doi: 10.1534/genetics.117.300482. Genetics. 2018. PMID: 29301949 Free PMC article. Review.
-
Identification of Suppressors of top-2 Embryonic Lethality in Caenorhabditis elegans.G3 (Bethesda). 2020 Apr 9;10(4):1183-1191. doi: 10.1534/g3.119.400927. G3 (Bethesda). 2020. PMID: 32086248 Free PMC article.
-
Meiosis.WormBook. 2017 May 4;2017:1-43. doi: 10.1895/wormbook.1.178.1. WormBook. 2017. PMID: 26694509 Free PMC article. Review.
Cited by
-
Nutrient Deficiencies Impact on the Cellular and Metabolic Responses of Saxitoxin Producing Alexandrium minutum: A Transcriptomic Perspective.Mar Drugs. 2023 Sep 18;21(9):497. doi: 10.3390/md21090497. Mar Drugs. 2023. PMID: 37755110 Free PMC article.
-
Topoisomerases Modulate the Timing of Meiotic DNA Breakage and Chromosome Morphogenesis in Saccharomyces cerevisiae.Genetics. 2020 May;215(1):59-73. doi: 10.1534/genetics.120.303060. Epub 2020 Mar 9. Genetics. 2020. PMID: 32152049 Free PMC article.
-
Harnessing the power of genetics: fast forward genetics in Caenorhabditis elegans.Mol Genet Genomics. 2021 Jan;296(1):1-20. doi: 10.1007/s00438-020-01721-6. Epub 2020 Sep 4. Mol Genet Genomics. 2021. PMID: 32888055 Review.
-
The demethylase NMAD-1 regulates DNA replication and repair in the Caenorhabditis elegans germline.PLoS Genet. 2019 Jul 8;15(7):e1008252. doi: 10.1371/journal.pgen.1008252. eCollection 2019 Jul. PLoS Genet. 2019. PMID: 31283754 Free PMC article.
-
Evaluating alignment and variant-calling software for mutation identification in C. elegans by whole-genome sequencing.PLoS One. 2017 Mar 23;12(3):e0174446. doi: 10.1371/journal.pone.0174446. eCollection 2017. PLoS One. 2017. PMID: 28333980 Free PMC article.
References
-
- Albertson D. G., Thomson J. N., 1993. Segregation of holocentric chromosomes at meiosis in the nematode, Caenorhabditis elegans. Chromosome Res. 1: 15–26. - PubMed
-
- Bhat M. A., Philp A. V., Glover D. M., Bellen H. J., 1996. Chromatid segregation at anaphase requires the barren product, a novel chromosome-associated protein that interacts with Topoisomerase II. Cell 87: 1103–1114. - PubMed
-
- Braun R. E., 2001. Packaging paternal chromosomes with protamine. Nat. Genet. 28: 10–12. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

