Epstein-Barr virus microRNAs reduce immune surveillance by virus-specific CD8+ T cells
- PMID: 27698133
- PMCID: PMC5081573
- DOI: 10.1073/pnas.1605884113
Epstein-Barr virus microRNAs reduce immune surveillance by virus-specific CD8+ T cells
Abstract
Infection with Epstein-Barr virus (EBV) affects most humans worldwide and persists life-long in the presence of robust virus-specific T-cell responses. In both immunocompromised and some immunocompetent people, EBV causes several cancers and lymphoproliferative diseases. EBV transforms B cells in vitro and encodes at least 44 microRNAs (miRNAs), most of which are expressed in EBV-transformed B cells, but their functions are largely unknown. Recently, we showed that EBV miRNAs inhibit CD4+ T-cell responses to infected B cells by targeting IL-12, MHC class II, and lysosomal proteases. Here we investigated whether EBV miRNAs also counteract surveillance by CD8+ T cells. We have found that EBV miRNAs strongly inhibit recognition and killing of infected B cells by EBV-specific CD8+ T cells through multiple mechanisms. EBV miRNAs directly target the peptide transporter subunit TAP2 and reduce levels of the TAP1 subunit, MHC class I molecules, and EBNA1, a protein expressed in most forms of EBV latency and a target of EBV-specific CD8+ T cells. Moreover, miRNA-mediated down-regulation of the cytokine IL-12 decreases the recognition of infected cells by EBV-specific CD8+ T cells. Thus, EBV miRNAs use multiple, distinct pathways, allowing the virus to evade surveillance not only by CD4+ but also by antiviral CD8+ T cells.
Keywords: CD8 T cells; adaptive immunity; herpesvirus; immune evasion; microRNA.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

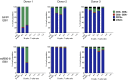

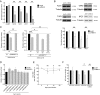
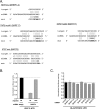

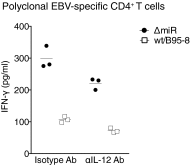
Similar articles
-
Epstein-Barr virus evasion of CD8(+) and CD4(+) T cell immunity via concerted actions of multiple gene products.Semin Cancer Biol. 2008 Dec;18(6):397-408. doi: 10.1016/j.semcancer.2008.10.008. Epub 2008 Oct 25. Semin Cancer Biol. 2008. PMID: 18977445 Review.
-
MicroRNAs of Epstein-Barr Virus Attenuate T-Cell-Mediated Immune Control In Vivo.mBio. 2019 Jan 15;10(1):e01941-18. doi: 10.1128/mBio.01941-18. mBio. 2019. PMID: 30647153 Free PMC article.
-
Multiple Viral microRNAs Regulate Interferon Release and Signaling Early during Infection with Epstein-Barr Virus.mBio. 2021 Mar 30;12(2):e03440-20. doi: 10.1128/mBio.03440-20. mBio. 2021. PMID: 33785626 Free PMC article.
-
Stage-specific inhibition of MHC class I presentation by the Epstein-Barr virus BNLF2a protein during virus lytic cycle.PLoS Pathog. 2009 Jun;5(6):e1000490. doi: 10.1371/journal.ppat.1000490. Epub 2009 Jun 26. PLoS Pathog. 2009. PMID: 19557156 Free PMC article.
-
Role of Viral and Host microRNAs in Immune Regulation of Epstein-Barr Virus-Associated Diseases.Front Immunol. 2020 Mar 3;11:367. doi: 10.3389/fimmu.2020.00367. eCollection 2020. Front Immunol. 2020. PMID: 32194570 Free PMC article. Review.
Cited by
-
Epstein-Barr virus: the mastermind of immune chaos.Front Immunol. 2024 Feb 7;15:1297994. doi: 10.3389/fimmu.2024.1297994. eCollection 2024. Front Immunol. 2024. PMID: 38384471 Free PMC article. Review.
-
High MHC-II expression in Epstein-Barr virus-associated gastric cancers suggests that tumor cells serve an important role in antigen presentation.Sci Rep. 2020 Sep 8;10(1):14786. doi: 10.1038/s41598-020-71775-4. Sci Rep. 2020. PMID: 32901107 Free PMC article.
-
Novel approach to identify putative Epstein-Barr-virus microRNAs regulating host cell genes with relevance in tumor biology and immunology.Oncoimmunology. 2022 May 1;11(1):2070338. doi: 10.1080/2162402X.2022.2070338. eCollection 2022. Oncoimmunology. 2022. PMID: 35529676 Free PMC article.
-
Current status and advances of immunotherapy in nasopharyngeal carcinoma.Ther Adv Med Oncol. 2022 May 7;14:17588359221096214. doi: 10.1177/17588359221096214. eCollection 2022. Ther Adv Med Oncol. 2022. PMID: 35547095 Free PMC article. Review.
-
High Levels of Class I Major Histocompatibility Complex mRNA Are Present in Epstein-Barr Virus-Associated Gastric Adenocarcinomas.Cells. 2020 Feb 21;9(2):499. doi: 10.3390/cells9020499. Cells. 2020. PMID: 32098275 Free PMC article.
References
-
- van Baarle D, et al. Dysfunctional Epstein-Barr virus (EBV)-specific CD8(+) T lymphocytes and increased EBV load in HIV-1 infected individuals progressing to AIDS-related non-Hodgkin lymphoma. Blood. 2001;98(1):146–155. - PubMed
-
- Smets F, et al. Ratio between Epstein-Barr viral load and anti-Epstein-Barr virus specific T-cell response as a predictive marker of posttransplant lymphoproliferative disease. Transplantation. 2002;73(10):1603–1610. - PubMed
-
- Moosmann A, et al. Effective and long-term control of EBV PTLD after transfer of peptide-selected T cells. Blood. 2010;115(14):2960–2970. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

