Functional interplay between Mediator and TFIIB in preinitiation complex assembly in relation to promoter architecture
- PMID: 27688401
- PMCID: PMC5066617
- DOI: 10.1101/gad.285775.116
Functional interplay between Mediator and TFIIB in preinitiation complex assembly in relation to promoter architecture
Abstract
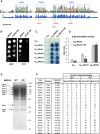
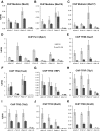
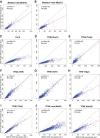
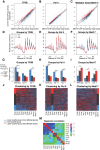
Mediator is a large coregulator complex conserved from yeast to humans and involved in many human diseases, including cancers. Together with general transcription factors, it stimulates preinitiation complex (PIC) formation and activates RNA polymerase II (Pol II) transcription. In this study, we analyzed how Mediator acts in PIC assembly using in vivo, in vitro, and in silico approaches. We revealed an essential function of the Mediator middle module exerted through its Med10 subunit, implicating a key interaction between Mediator and TFIIB. We showed that this Mediator-TFIIB link has a global role on PIC assembly genome-wide. Moreover, the amplitude of Mediator's effect on PIC formation is gene-dependent and is related to the promoter architecture in terms of TATA elements, nucleosome occupancy, and dynamics. This study thus provides mechanistic insights into the coordinated function of Mediator and TFIIB in PIC assembly in different chromatin contexts.
Keywords: Mediator; RNA polymerase II transcription; Saccharomyces cerevisiae; TFIIB; preinitiation complex; promoter architecture.
© 2016 Eychenne et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


Similar articles
-
Mediator, TATA-binding protein, and RNA polymerase II contribute to low histone occupancy at active gene promoters in yeast.J Biol Chem. 2014 May 23;289(21):14981-95. doi: 10.1074/jbc.M113.529354. Epub 2014 Apr 11. J Biol Chem. 2014. PMID: 24727477 Free PMC article.
-
Mediator independently orchestrates multiple steps of preinitiation complex assembly in vivo.Nucleic Acids Res. 2015 Oct 30;43(19):9214-31. doi: 10.1093/nar/gkv782. Epub 2015 Aug 3. Nucleic Acids Res. 2015. PMID: 26240385 Free PMC article.
-
Spatiotemporal coordination of transcription preinitiation complex assembly in live cells.Mol Cell. 2021 Sep 2;81(17):3560-3575.e6. doi: 10.1016/j.molcel.2021.07.022. Epub 2021 Aug 9. Mol Cell. 2021. PMID: 34375585 Free PMC article.
-
Toward understanding of the mechanisms of Mediator function in vivo: Focus on the preinitiation complex assembly.Transcription. 2017;8(5):328-342. doi: 10.1080/21541264.2017.1329000. Epub 2017 Aug 25. Transcription. 2017. PMID: 28841352 Free PMC article. Review.
-
More pieces to the puzzle: recent structural insights into class II transcription initiation.Curr Opin Struct Biol. 2014 Feb;24:91-7. doi: 10.1016/j.sbi.2013.12.005. Epub 2014 Jan 16. Curr Opin Struct Biol. 2014. PMID: 24440461 Review.
Cited by
-
The Mediator Complex: A Central Coordinator of Plant Adaptive Responses to Environmental Stresses.Int J Mol Sci. 2022 May 31;23(11):6170. doi: 10.3390/ijms23116170. Int J Mol Sci. 2022. PMID: 35682844 Free PMC article. Review.
-
Yeast Mediator facilitates transcription initiation at most promoters via a Tail-independent mechanism.Mol Cell. 2022 Nov 3;82(21):4033-4048.e7. doi: 10.1016/j.molcel.2022.09.016. Epub 2022 Oct 7. Mol Cell. 2022. PMID: 36208626 Free PMC article.
-
Mediator Is Essential for Small Nuclear and Nucleolar RNA Transcription in Yeast.Mol Cell Biol. 2018 Nov 28;38(24):e00296-18. doi: 10.1128/MCB.00296-18. Print 2018 Dec 15. Mol Cell Biol. 2018. PMID: 30275344 Free PMC article.
-
Eukaryotic core promoters and the functional basis of transcription initiation.Nat Rev Mol Cell Biol. 2018 Oct;19(10):621-637. doi: 10.1038/s41580-018-0028-8. Nat Rev Mol Cell Biol. 2018. PMID: 29946135 Free PMC article. Review.
-
Transcription regulation by the Mediator complex.Nat Rev Mol Cell Biol. 2018 Apr;19(4):262-274. doi: 10.1038/nrm.2017.115. Epub 2017 Dec 6. Nat Rev Mol Cell Biol. 2018. PMID: 29209056 Review.
References
-
- Baek HJ, Kang YK, Roeder RG. 2006. Human Mediator enhances basal transcription by facilitating recruitment of transcription factor IIB during preinitiation complex assembly. J Biol Chem 281: 15172–15181. - PubMed
-
- Buratowski S, Hahn S, Guarente L, Sharp PA. 1989. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell 56: 549–561. - PubMed
-
- Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. 2007. Dynamics of replication-independent histone turnover in budding yeast. Science 315: 1405–1408. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
