E3 Ubiquitin Ligase RLIM Negatively Regulates c-Myc Transcriptional Activity and Restrains Cell Proliferation
- PMID: 27684546
- PMCID: PMC5042457
- DOI: 10.1371/journal.pone.0164086
E3 Ubiquitin Ligase RLIM Negatively Regulates c-Myc Transcriptional Activity and Restrains Cell Proliferation
Abstract
RNF12/RLIM is a RING domain-containing E3 ubiquitin ligase whose function has only begun to be elucidated recently. Although RLIM was reported to play important roles in some biological processes such as imprinted X-chromosome inactivation and regulation of TGF-β pathway etc., other functions of RLIM are largely unknown. Here, we identified RLIM as a novel E3 ubiquitin ligase for c-Myc, one of the most frequently deregulated oncoproteins in human cancers. RLIM associates with c-Myc in vivo and in vitro independently of the E3 ligase activity of RLIM. Moreover, RLIM promotes the polyubiquitination of c-Myc protein independently of Ser62 and Thr58 phosphorylation of c-Myc. However, RLIM-mediated ubiquitination does not affect c-Myc stability. Instead, RLIM inhibits the transcriptional activity of c-Myc through which RLIM restrains cell proliferation. Our results suggest that RLIM may function as a tumor suppressor by controlling the activity of c-Myc oncoprotein.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

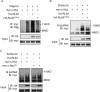
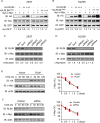
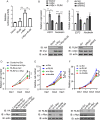
Similar articles
-
Kaposi's Sarcoma-Associated Herpesvirus LANA Modulates the Stability of the E3 Ubiquitin Ligase RLIM.J Virol. 2020 Feb 14;94(5):e01578-19. doi: 10.1128/JVI.01578-19. Print 2020 Feb 14. J Virol. 2020. PMID: 31801865 Free PMC article.
-
Pathogenic variants in E3 ubiquitin ligase RLIM/RNF12 lead to a syndromic X-linked intellectual disability and behavior disorder.Mol Psychiatry. 2019 Nov;24(11):1748-1768. doi: 10.1038/s41380-018-0065-x. Epub 2018 May 4. Mol Psychiatry. 2019. PMID: 29728705
-
Rlim/Rnf12, Rex1, and X Chromosome Inactivation.Front Cell Dev Biol. 2019 Oct 31;7:258. doi: 10.3389/fcell.2019.00258. eCollection 2019. Front Cell Dev Biol. 2019. PMID: 31737626 Free PMC article.
-
Balance of Yin and Yang: ubiquitylation-mediated regulation of p53 and c-Myc.Neoplasia. 2006 Aug;8(8):630-44. doi: 10.1593/neo.06334. Neoplasia. 2006. PMID: 16925946 Free PMC article. Review.
-
Targeting the MYC Ubiquitination-Proteasome Degradation Pathway for Cancer Therapy.Front Oncol. 2021 Jun 11;11:679445. doi: 10.3389/fonc.2021.679445. eCollection 2021. Front Oncol. 2021. PMID: 34178666 Free PMC article. Review.
Cited by
-
Aspirin regulation of c-myc and cyclinD1 proteins to overcome tamoxifen resistance in estrogen receptor-positive breast cancer cells.Oncotarget. 2017 May 2;8(18):30252-30264. doi: 10.18632/oncotarget.16325. Oncotarget. 2017. PMID: 28415819 Free PMC article.
-
RNF12 is regulated by AKT phosphorylation and promotes TGF-β driven breast cancer metastasis.Cell Death Dis. 2022 Jan 10;13(1):44. doi: 10.1038/s41419-021-04493-y. Cell Death Dis. 2022. PMID: 35013159 Free PMC article.
-
Activity-based probe profiling of RNF12 E3 ubiquitin ligase function in Tonne-Kalscheuer syndrome.Life Sci Alliance. 2022 Jun 28;5(11):e202101248. doi: 10.26508/lsa.202101248. Print 2022 Nov. Life Sci Alliance. 2022. PMID: 35764390 Free PMC article.
-
Proteasomal Degradation of Zn-Dependent Hdacs: The E3-Ligases Implicated and the Designed Protacs That Enable Degradation.Molecules. 2021 Sep 15;26(18):5606. doi: 10.3390/molecules26185606. Molecules. 2021. PMID: 34577077 Free PMC article. Review.
-
RLIM suppresses hepatocellular carcinogenesis by up-regulating p15 and p21.Oncotarget. 2017 Sep 15;8(47):83075-83087. doi: 10.18632/oncotarget.20904. eCollection 2017 Oct 10. Oncotarget. 2017. PMID: 29137325 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

