Purification of the subcellular compartment in which exogenous antigens undergo endoplasmic reticulum-associated degradation from dendritic cells
- PMID: 27656684
- PMCID: PMC5021789
- DOI: 10.1016/j.heliyon.2016.e00151
Purification of the subcellular compartment in which exogenous antigens undergo endoplasmic reticulum-associated degradation from dendritic cells
Abstract
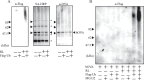
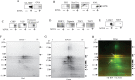
Dendritic cells (DCs) are capable of processing and presenting exogenous antigens using MHC class I molecules. This pathway is called antigen cross-presentation and plays an important role in the stimulation of naïve CD8(+) T cells for infectious and tumor immunity. Our previous studies in DC2.4 cells and bone marrow-derived DCs revealed that exogenously added ovalbumin (OVA) is processed through endoplasmic reticulum (ER)-associated degradation (ERAD) for cross-presentation. In this study, we aimed to further confirm these results by purification of the subcellular compartment in which exogenous antigens undergo ERAD from homogenates of DC2.4 cells pretreated with biotinylated OVA (bOVA). bOVA-containing vesicles were purified using streptavidin (SA)-magnetic beads from cell homogenates and were found to contain ER chaperones and ERAD components together with proteins for antigen presentation. In purified microsomes, bOVA was retained in membranous fractions and degraded by the ubiquitin proteasome system in presence reticulocyte lysates and ATP. These results strongly suggested that DCs processed and degraded exogenous antigens through ERAD for cross-presentation in this purified subcellular compartment.
Keywords: Cell biology; Immunology.
Figures



Similar articles
-
Defining the optimum strategy for identifying adults and children with coeliac disease: systematic review and economic modelling.Health Technol Assess. 2022 Oct;26(44):1-310. doi: 10.3310/ZUCE8371. Health Technol Assess. 2022. PMID: 36321689 Free PMC article.
-
Electronic cigarettes for smoking cessation.Cochrane Database Syst Rev. 2024 Jan 8;1(1):CD010216. doi: 10.1002/14651858.CD010216.pub8. Cochrane Database Syst Rev. 2024. PMID: 38189560 Free PMC article.
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
Impact of residual disease as a prognostic factor for survival in women with advanced epithelial ovarian cancer after primary surgery.Cochrane Database Syst Rev. 2022 Sep 26;9(9):CD015048. doi: 10.1002/14651858.CD015048.pub2. Cochrane Database Syst Rev. 2022. PMID: 36161421 Free PMC article. Review.
Cited by
-
Ultra-low volume intradermal administration of radiation-attenuated sporozoites with the glycolipid adjuvant 7DW8-5 completely protects mice against malaria.Sci Rep. 2024 Feb 4;14(1):2881. doi: 10.1038/s41598-024-53118-9. Sci Rep. 2024. PMID: 38311678 Free PMC article.
-
Distinct Subcellular Compartments of Dendritic Cells Used for Cross-Presentation.Int J Mol Sci. 2019 Nov 9;20(22):5606. doi: 10.3390/ijms20225606. Int J Mol Sci. 2019. PMID: 31717517 Free PMC article. Review.
-
Purification of the Membrane Compartment for Endoplasmic Reticulum-associated Degradation of Exogenous Antigens in Cross-presentation.J Vis Exp. 2017 Aug 21;(126):55949. doi: 10.3791/55949. J Vis Exp. 2017. PMID: 28872140 Free PMC article.
-
Endoplasmic Reticulum-Associated Degradation-Dependent Processing in Cross-Presentation and Its Potential for Dendritic Cell Vaccinations: A Review.Pharmaceutics. 2020 Feb 13;12(2):153. doi: 10.3390/pharmaceutics12020153. Pharmaceutics. 2020. PMID: 32070016 Free PMC article. Review.
References
-
- Janeway C., Travers P., Walport M., Shlomchik M. 5th edn. Garland Press; New York: 2001. Immunobiology: The Immune System in Health and Disease.
-
- Yewdell J., Anton L.C., Bacik I., Schubert U., Snyder H.L., Bennink J.R. Generating MHC class I ligands from viral gene products. Immunol. Rev. 1999;172:97–108. - PubMed
-
- Huang A.Y., Golumbek P., Ahmadzadeh M., Jaffee E., Pardoll D., Levitsky H. Role of bone marrow-derived cells in presenting MHC class I-restricted tumor antigens. Science. 1994;264:961–965. - PubMed
-
- Banchereau J., Steinman R.M. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. - PubMed
-
- Bevan M.J., Minor H. Antigens introduced on H-2 different stimulating cells cross-react at the cytotoxic T cell level during in vivo priming. J. Immunol. 1976;117:2233–2238. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

