Influence of Human p53 on Plant Development
- PMID: 27648563
- PMCID: PMC5029891
- DOI: 10.1371/journal.pone.0162840
Influence of Human p53 on Plant Development
Abstract
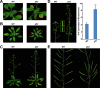
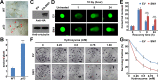
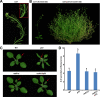
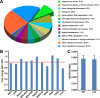
Mammalian p53 is a super tumor suppressor and plays a key role in guarding genome from DNA damage. However, p53 has not been found in plants which do not bear cancer although they constantly expose to ionizing radiation of ultraviolet light. Here we introduced p53 into the model plant Arabidopsis and examined p53-conferred phenotype in plant. Most strikingly, p53 caused early senescence and fasciation. In plants, fasciation has been shown as a result of the elevated homologous DNA recombination. Consistently, a reporter with overlapping segments of the GUS gene (1445) showed that the frequency of homologous recombination was highly induced in p53-transgenic plants. In contrast to p53, SUPPRESSOR OF NPR1-1 INDUCIBLE 1 (SNI1), as a negative regulator of homologous recombination in plants, is not present in mammals. Comet assay and clonogenic survival assay demonstrated that SNI1 inhibited DNA damage repair caused by either ionizing radiation or hydroxyurea in human osteosarcoma U2OS cancer cells. RAD51D is a recombinase in homologous recombination and functions downstream of SNI1 in plants. Interestingly, p53 rendered the sni1 mutants madly branching of inflorescence, a phenotype of fasciation, whereas rad51d mutant fully suppressed the p53-induced phenotype, indicating that human p53 action in plant is mediated by the SNI1-RAD51D signaling pathway. The reciprocal species-swap tests of p53 and SNI1 in human and Arabidopsis manifest that these species-specific proteins play a common role in homologous recombination across kingdoms of animals and plants.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Arabidopsis SNI1 and RAD51D regulate both gene transcription and DNA recombination during the defense response.Proc Natl Acad Sci U S A. 2007 Mar 6;104(10):4223-7. doi: 10.1073/pnas.0609357104. Epub 2007 Feb 21. Proc Natl Acad Sci U S A. 2007. PMID: 17360504 Free PMC article.
-
DNA repair proteins are directly involved in regulation of gene expression during plant immune response.Cell Host Microbe. 2011 Feb 17;9(2):115-24. doi: 10.1016/j.chom.2011.01.011. Cell Host Microbe. 2011. PMID: 21320694
-
Negative regulator of E2F transcription factors links cell cycle checkpoint and DNA damage repair.Proc Natl Acad Sci U S A. 2018 Apr 17;115(16):E3837-E3845. doi: 10.1073/pnas.1720094115. Epub 2018 Apr 2. Proc Natl Acad Sci U S A. 2018. PMID: 29610335 Free PMC article.
-
The role of SOG1, a plant-specific transcriptional regulator, in the DNA damage response.Plant Signal Behav. 2014;9(4):e28889. doi: 10.4161/psb.28889. Plant Signal Behav. 2014. PMID: 24736489 Free PMC article. Review.
-
Reevaluation of the reliability and usefulness of the somatic homologous recombination reporter lines.Plant Cell. 2012 Nov;24(11):4314-23. doi: 10.1105/tpc.112.100404. Epub 2012 Nov 9. Plant Cell. 2012. PMID: 23144181 Free PMC article. Review.
Cited by
-
The foundational framework of tumors: Gametogenesis, p53, and cancer.Semin Cancer Biol. 2022 Jun;81:193-205. doi: 10.1016/j.semcancer.2021.04.018. Epub 2021 Apr 30. Semin Cancer Biol. 2022. PMID: 33940178 Free PMC article. Review.
-
The Multiple Functions of the Nucleolus in Plant Development, Disease and Stress Responses.Front Plant Sci. 2018 Feb 9;9:132. doi: 10.3389/fpls.2018.00132. eCollection 2018. Front Plant Sci. 2018. PMID: 29479362 Free PMC article. Review.
-
DNA Damage Repair System in Plants: A Worldwide Research Update.Genes (Basel). 2017 Oct 30;8(11):299. doi: 10.3390/genes8110299. Genes (Basel). 2017. PMID: 29084140 Free PMC article. Review.
References
-
- Marchenko ND, Moll UM. The role of ubiquitination in the direct mitochondrial death program of p53. Cell cycle. 2007; 6(14): 1718–23. . - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

