Structure of the unliganded form of the proprotein convertase furin suggests activation by a substrate-induced mechanism
- PMID: 27647913
- PMCID: PMC5056075
- DOI: 10.1073/pnas.1613630113
Structure of the unliganded form of the proprotein convertase furin suggests activation by a substrate-induced mechanism
Abstract
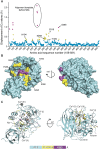
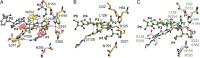
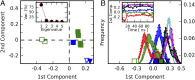
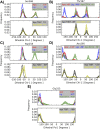
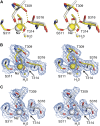
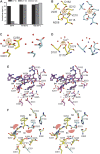
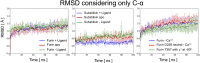
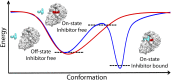
Proprotein convertases (PCs) are highly specific proteases required for the proteolytic modification of many secreted proteins. An unbalanced activity of these enzymes is connected to pathologies like cancer, atherosclerosis, hypercholesterolaemia, and infectious diseases. Novel protein crystallographic structures of the prototypical PC family member furin in different functional states were determined to 1.8-2.0 Å. These, together with biochemical data and modeling by molecular dynamics calculations, suggest essential elements underlying its unusually high substrate specificity. Furin shows a complex activation mechanism and exists in at least four defined states: (i) the "off state," incompatible with substrate binding as seen in the unliganded enzyme; (ii) the active "on state" seen in inhibitor-bound furin; and the respective (iii) calcium-free and (iv) calcium-bound forms. The transition from the off to the on state is triggered by ligand binding at subsites S1 to S4 and appears to underlie the preferential recognition of the four-residue sequence motif of furin. The molecular dynamics simulations of the four structural states reflect the experimental observations in general and provide approximations of the respective stabilities. Ligation by calcium at the PC-specific binding site II influences the active-site geometry and determines the rotamer state of the oxyanion hole-forming Asn295, and thus adds a second level of the activity modulation of furin. The described crystal forms and the observations of different defined functional states may foster the development of new tools and strategies for pharmacological intervention targeting furin.
Keywords: activation; conformational transition; serine-protease; specificity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
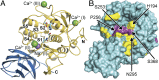


Similar articles
-
OFF-State-Specific Inhibition of the Proprotein Convertase Furin.ACS Chem Biol. 2021 Sep 17;16(9):1692-1700. doi: 10.1021/acschembio.1c00411. Epub 2021 Aug 20. ACS Chem Biol. 2021. PMID: 34415722 Free PMC article.
-
Structural Studies Revealed Active Site Distortions of Human Furin by a Small Molecule Inhibitor.ACS Chem Biol. 2017 May 19;12(5):1211-1216. doi: 10.1021/acschembio.6b01110. Epub 2017 Apr 17. ACS Chem Biol. 2017. PMID: 28402100
-
X-ray Structures of the Proprotein Convertase Furin Bound with Substrate Analogue Inhibitors Reveal Substrate Specificity Determinants beyond the S4 Pocket.Biochemistry. 2018 Feb 13;57(6):925-934. doi: 10.1021/acs.biochem.7b01124. Epub 2018 Jan 26. Biochemistry. 2018. PMID: 29314830
-
The kindest cuts of all: crystal structures of Kex2 and furin reveal secrets of precursor processing.Trends Biochem Sci. 2004 Feb;29(2):80-7. doi: 10.1016/j.tibs.2003.12.006. Trends Biochem Sci. 2004. PMID: 15102434 Review.
-
[Furin and its biological role].Ukr Biokhim Zh (1999). 2007 Nov-Dec;79(6):5-18. Ukr Biokhim Zh (1999). 2007. PMID: 18712106 Review. Russian.
Cited by
-
Capturing a Crucial 'Disorder-to-Order Transition' at the Heart of the Coronavirus Molecular Pathology-Triggered by Highly Persistent, Interchangeable Salt-Bridges.Vaccines (Basel). 2022 Feb 16;10(2):301. doi: 10.3390/vaccines10020301. Vaccines (Basel). 2022. PMID: 35214759 Free PMC article.
-
Spread of Gamma (P.1) Sub-Lineages Carrying Spike Mutations Close to the Furin Cleavage Site and Deletions in the N-Terminal Domain Drives Ongoing Transmission of SARS-CoV-2 in Amazonas, Brazil.Microbiol Spectr. 2022 Feb 23;10(1):e0236621. doi: 10.1128/spectrum.02366-21. Epub 2022 Feb 23. Microbiol Spectr. 2022. PMID: 35196783 Free PMC article.
-
Dichlorophenylpyridine-Based Molecules Inhibit Furin through an Induced-Fit Mechanism.ACS Chem Biol. 2022 Apr 15;17(4):816-821. doi: 10.1021/acschembio.2c00103. Epub 2022 Apr 4. ACS Chem Biol. 2022. PMID: 35377598 Free PMC article.
-
Structure-Based Macrocyclization of Substrate Analogue NS2B-NS3 Protease Inhibitors of Zika, West Nile and Dengue viruses.ChemMedChem. 2020 Aug 5;15(15):1439-1452. doi: 10.1002/cmdc.202000237. Epub 2020 Jun 30. ChemMedChem. 2020. PMID: 32501637 Free PMC article.
-
Comparison of clinically approved molecules on SARS-CoV-2 drug target proteins: a molecular docking study.Turk J Chem. 2021 Feb 17;45(1):35-41. doi: 10.3906/kim-2008-35. eCollection 2021. Turk J Chem. 2021. PMID: 33679150 Free PMC article.
References
-
- Artenstein AW, Opal SM. Proprotein convertases in health and disease. N Engl J Med. 2011;365(26):2507–2518. - PubMed
-
- Seidah NG, Prat A. The biology and therapeutic targeting of the proprotein convertases. Nat Rev Drug Discov. 2012;11(5):367–383. - PubMed
-
- Garten W, et al. Processing of viral glycoproteins by the subtilisin-like endoprotease furin and its inhibition by specific peptidylchloroalkylketones. Biochimie. 1994;76(3-4):217–225. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

