Sequence analysis of the Hsp70 family in moss and evaluation of their functions in abiotic stress responses
- PMID: 27644410
- PMCID: PMC5028893
- DOI: 10.1038/srep33650
Sequence analysis of the Hsp70 family in moss and evaluation of their functions in abiotic stress responses
Abstract
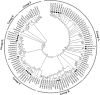
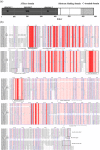
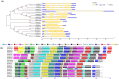
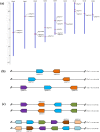
The 70-kD heat shock proteins (Hsp70s) are highly conserved molecular chaperones that play essential roles in cellular processes including abiotic stress responses. Physcomitrella patens serves as a representative of the first terrestrial plants and can recover from serious dehydration. To assess the possible relationship between P. patens Hsp70s and dehydration tolerance, we analyzed the P. patens genome and found at least 21 genes encoding Hsp70s. Gene structure and motif composition were relatively conserved in each subfamily. The intron-exon structure of PpcpHsp70-2 was different from that of other PpcpHsp70s; this gene exhibits several forms of intron retention, indicating that introns may play important roles in regulating gene expression. We observed expansion of Hsp70s in P. patens, which may reflect adaptations related to development and dehydration tolerance, and results mainly from tandem and segmental duplications. Expression profiles of rice, Arabidopsis and P. patens Hsp70 genes revealed that more than half of the Hsp70 genes were responsive to ABA, salt and drought. The presence of overrepresented cis-elements (DOFCOREZM and GCCCORE) among stress-responsive Hsp70s suggests that they share a common regulatory pathway. Moss plants overexpressing PpcpHsp70-2 showed salt and dehydration tolerance, further supporting a role in adaptation to land. This work highlights directions for future functional analyses of Hsp70s.
Figures


Similar articles
-
Genome-Wide Identification and Expression Analysis of Heat Shock Protein 70 (HSP70) Gene Family in Pumpkin (Cucurbita moschata) Rootstock under Drought Stress Suggested the Potential Role of these Chaperones in Stress Tolerance.Int J Mol Sci. 2022 Feb 8;23(3):1918. doi: 10.3390/ijms23031918. Int J Mol Sci. 2022. PMID: 35163839 Free PMC article.
-
Genome-wide survey of heat shock factors and heat shock protein 70s and their regulatory network under abiotic stresses in Brachypodium distachyon.PLoS One. 2017 Jul 6;12(7):e0180352. doi: 10.1371/journal.pone.0180352. eCollection 2017. PLoS One. 2017. PMID: 28683139 Free PMC article.
-
Genome-wide identification and characterization of Hsp70 gene family in Nicotiana tabacum.Mol Biol Rep. 2019 Apr;46(2):1941-1954. doi: 10.1007/s11033-019-04644-7. Epub 2019 Feb 1. Mol Biol Rep. 2019. PMID: 30710231
-
Roles of Hsp70s in Stress Responses of Microorganisms, Plants, and Animals.Biomed Res Int. 2015;2015:510319. doi: 10.1155/2015/510319. Epub 2015 Nov 16. Biomed Res Int. 2015. PMID: 26649306 Free PMC article. Review.
-
Moss transcription factors regulating development and defense responses to stress.J Exp Bot. 2022 Jul 16;73(13):4546-4561. doi: 10.1093/jxb/erac055. J Exp Bot. 2022. PMID: 35167679 Review.
Cited by
-
Comparative proteomic analysis reveals that the Heterosis of two maize hybrids is related to enhancement of stress response and photosynthesis respectively.BMC Plant Biol. 2021 Jan 9;21(1):34. doi: 10.1186/s12870-020-02806-5. BMC Plant Biol. 2021. PMID: 33422018 Free PMC article.
-
Evolutionary analysis and expression profiling of the HSP70 gene family in response to abiotic stresses in tomato (Solanum lycopersicum).Sci Prog. 2023 Jan-Mar;106(1):368504221148843. doi: 10.1177/00368504221148843. Sci Prog. 2023. PMID: 36650980 Free PMC article.
-
Translocation of Drought-Responsive Proteins from the Chloroplasts.Cells. 2020 Jan 20;9(1):259. doi: 10.3390/cells9010259. Cells. 2020. PMID: 31968705 Free PMC article.
-
Different Roles of Heat Shock Proteins (70 kDa) During Abiotic Stresses in Barley (Hordeum vulgare) Genotypes.Plants (Basel). 2019 Jul 26;8(8):248. doi: 10.3390/plants8080248. Plants (Basel). 2019. PMID: 31357401 Free PMC article.
-
Cold Acclimation Improves the Desiccation Stress Resilience of Polar Strains of Klebsormidium (Streptophyta).Front Microbiol. 2019 Aug 6;10:1730. doi: 10.3389/fmicb.2019.01730. eCollection 2019. Front Microbiol. 2019. PMID: 31447802 Free PMC article.
References
-
- Feder M. E. & Hofmann G. E. Heat-shock proteins, molecular chaperones, and the stress response: Evolutionary and ecological physiology. Annu Rev Physiol 61, 243–282 (1999). - PubMed
-
- Georgopoulos C. & Welch W. J. Role of the major heat-shock proteins as molecular chaperones. Annu Rev Cell Biol 9, 601–634 (1993). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

