Mechanism of Assembly and Cooperativity of Homomeric and Heteromeric Metabotropic Glutamate Receptors
- PMID: 27641494
- PMCID: PMC5053906
- DOI: 10.1016/j.neuron.2016.08.036
Mechanism of Assembly and Cooperativity of Homomeric and Heteromeric Metabotropic Glutamate Receptors
Abstract
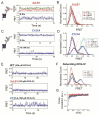
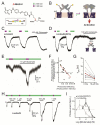
G protein-coupled receptors (GPCRs) mediate cellular responses to a wide variety of extracellular stimuli. GPCR dimerization may expand signaling diversity and tune functionality, but little is known about the mechanisms of subunit assembly and interaction or the signaling properties of heteromers. Using single-molecule subunit counting on class C metabotropic glutamate receptors (mGluRs), we map dimerization determinants and define a heterodimerization profile. Intersubunit fluorescence resonance energy transfer measurements reveal that interactions between ligand-binding domains control the conformational rearrangements underlying receptor activation. Selective liganding with photoswitchable tethered agonists conjugated to one or both subunits of covalently linked mGluR2 homodimers reveals that receptor activation is highly cooperative. Strikingly, this cooperativity is asymmetric in mGluR2/mGluR3 heterodimers. Our results lead to a model of cooperative activation of mGluRs that provides a framework for understanding how class C GPCRs couple extracellular binding to dimer reorganization and G protein activation.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures

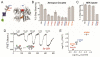
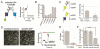
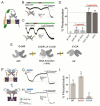
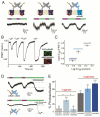

Similar articles
-
Allosteric control of an asymmetric transduction in a G protein-coupled receptor heterodimer.Elife. 2017 Aug 10;6:e26985. doi: 10.7554/eLife.26985. Elife. 2017. PMID: 28829739 Free PMC article.
-
Activation of a dimeric metabotropic glutamate receptor by intersubunit rearrangement.J Biol Chem. 2007 Nov 9;282(45):33000-8. doi: 10.1074/jbc.M702542200. Epub 2007 Sep 13. J Biol Chem. 2007. PMID: 17855348
-
Differences in interactions between transmembrane domains tune the activation of metabotropic glutamate receptors.Elife. 2021 Apr 21;10:e67027. doi: 10.7554/eLife.67027. Elife. 2021. PMID: 33880992 Free PMC article.
-
Structural and Biophysical Mechanisms of Class C G Protein-Coupled Receptor Function.Trends Biochem Sci. 2020 Dec;45(12):1049-1064. doi: 10.1016/j.tibs.2020.07.008. Epub 2020 Aug 26. Trends Biochem Sci. 2020. PMID: 32861513 Free PMC article. Review.
-
Novel functions for subtypes of metabotropic glutamate receptors.Neurochem Int. 1994 May;24(5):439-49. doi: 10.1016/0197-0186(94)90092-2. Neurochem Int. 1994. PMID: 7647699 Review.
Cited by
-
Optogenetic Techniques for Manipulating and Sensing G Protein-Coupled Receptor Signaling.Methods Mol Biol. 2020;2173:21-51. doi: 10.1007/978-1-0716-0755-8_2. Methods Mol Biol. 2020. PMID: 32651908 Free PMC article. Review.
-
Site-Specific Incorporation of Genetically Encoded Photo-Crosslinkers Locates the Heteromeric Interface of a GPCR Complex in Living Cells.Cell Chem Biol. 2020 Oct 15;27(10):1308-1317.e4. doi: 10.1016/j.chembiol.2020.07.006. Epub 2020 Jul 28. Cell Chem Biol. 2020. PMID: 32726588 Free PMC article.
-
Allosteric Modulation of GPCRs: New Insights and Potential Utility for Treatment of Schizophrenia and Other CNS Disorders.Neuron. 2017 May 3;94(3):431-446. doi: 10.1016/j.neuron.2017.03.016. Neuron. 2017. PMID: 28472649 Free PMC article. Review.
-
Pharmacological evidence for a metabotropic glutamate receptor heterodimer in neuronal cells.Elife. 2017 Jun 29;6:e25233. doi: 10.7554/eLife.25233. Elife. 2017. PMID: 28661401 Free PMC article.
-
Distinct beta-arrestin coupling and intracellular trafficking of metabotropic glutamate receptor homo- and heterodimers.Sci Adv. 2023 Dec 8;9(49):eadi8076. doi: 10.1126/sciadv.adi8076. Epub 2023 Dec 6. Sci Adv. 2023. PMID: 38055809 Free PMC article.
References
-
- Beqollari D, Kammermeier PJ. Venus fly trap domain of mGluR1 functions as a dominant negative against group I mGluR signaling. J Neurophysiol. 2010;104:439–448. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

