Analysis of genotype diversity and evolution of Dengue virus serotype 2 using complete genomes
- PMID: 27635316
- PMCID: PMC5012332
- DOI: 10.7717/peerj.2326
Analysis of genotype diversity and evolution of Dengue virus serotype 2 using complete genomes
Abstract
Background: Dengue is one of the most common arboviral diseases prevalent worldwide and is caused by Dengue viruses (genus Flavivirus, family Flaviviridae). There are four serotypes of Dengue Virus (DENV-1 to DENV-4), each of which is further subdivided into distinct genotypes. DENV-2 is frequently associated with severe dengue infections and epidemics. DENV-2 consists of six genotypes such as Asian/American, Asian I, Asian II, Cosmopolitan, American and sylvatic. Comparative genomic study was carried out to infer population structure of DENV-2 and to analyze the role of evolutionary and spatiotemporal factors in emergence of diversifying lineages.
Methods: Complete genome sequences of 990 strains of DENV-2 were analyzed using Bayesian-based population genetics and phylogenetic approaches to infer genetically distinct lineages. The role of spatiotemporal factors, genetic recombination and selection pressure in the evolution of DENV-2 is examined using the sequence-based bioinformatics approaches.
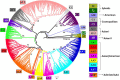
Results: DENV-2 genetic structure is complex and consists of fifteen subpopulations/lineages. The Asian/American genotype is observed to be diversified into seven lineages. The Asian I, Cosmopolitan and sylvatic genotypes were found to be subdivided into two lineages, each. The populations of American and Asian II genotypes were observed to be homogeneous. Significant evidence of episodic positive selection was observed in all the genes, except NS4A. Positive selection operational on a few codons in envelope gene confers antigenic and lineage diversity in the American strains of Asian/American genotype. Selection on codons of non-structural genes was observed to impact diversification of lineages in Asian I, cosmopolitan and sylvatic genotypes. Evidence of intra/inter-genotype recombination was obtained and the uncertainty in classification of recombinant strains was resolved using the population genetics approach.
Discussion: Complete genome-based analysis revealed that the worldwide population of DENV-2 strains is subdivided into fifteen lineages. The population structure of DENV-2 is spatiotemporal and is shaped by episodic positive selection and recombination. Intra-genotype diversity was observed in four genotypes (Asian/American, Asian I, cosmopolitan and sylvatic). Episodic positive selection on envelope and non-structural genes translates into antigenic diversity and appears to be responsible for emergence of strains/lineages in DENV-2 genotypes. Understanding of the genotype diversity and emerging lineages will be useful to design strategies for epidemiological surveillance and vaccine design.
Keywords: Bioinformatics; Comparative genomics; Dengue virus serotype 2; Evolution; Genetic structure; Genotype/genetic diversity; Molecular phylogeny; Population genetics; Recombination; Selection pressure.
Conflict of interest statement
The authors declare there are no competing interests.
Figures
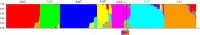
Similar articles
-
Genetic diversity and evolution of dengue virus serotype 3: A comparative genomics study.Infect Genet Evol. 2017 Apr;49:234-240. doi: 10.1016/j.meegid.2017.01.022. Epub 2017 Jan 23. Infect Genet Evol. 2017. PMID: 28126562
-
Population genomics of dengue virus serotype 4: insights into genetic structure and evolution.Arch Virol. 2016 Aug;161(8):2133-48. doi: 10.1007/s00705-016-2886-8. Epub 2016 May 12. Arch Virol. 2016. PMID: 27169727
-
Virus-like particle secretion and genotype-dependent immunogenicity of dengue virus serotype 2 DNA vaccine.J Virol. 2014 Sep;88(18):10813-30. doi: 10.1128/JVI.00810-14. Epub 2014 Jul 9. J Virol. 2014. PMID: 25008922 Free PMC article.
-
Comparative genomics and evolutionary analysis of dengue virus strains circulating in Pakistan.Virus Genes. 2024 Dec;60(6):603-620. doi: 10.1007/s11262-024-02100-8. Epub 2024 Aug 28. Virus Genes. 2024. PMID: 39198368
-
Recurrent Episodes of Some Mosquito-Borne Viral Diseases in Nigeria: A Systematic Review and Meta-Analysis.Pathogens. 2022 Oct 8;11(10):1162. doi: 10.3390/pathogens11101162. Pathogens. 2022. PMID: 36297219 Free PMC article. Review.
Cited by
-
Multiple arboviral infections during a DENV-2 outbreak in Solomon Islands.Trop Med Health. 2020 May 15;48:33. doi: 10.1186/s41182-020-00217-8. eCollection 2020. Trop Med Health. 2020. PMID: 32435149 Free PMC article.
-
Whole Genome Sequencing of DENV-2 isolated from Aedes aegypti mosquitoes in Esmeraldas, Ecuador. Genomic epidemiology of genotype III Southern Asian-American in the country.bioRxiv [Preprint]. 2024 Feb 8:2024.02.06.579255. doi: 10.1101/2024.02.06.579255. bioRxiv. 2024. PMID: 38370752 Free PMC article. Preprint.
-
Genetic Diversity of Dengue Virus in Clinical Specimens from Bangkok, Thailand, during 2018-2020: Co-Circulation of All Four Serotypes with Multiple Genotypes and/or Clades.Trop Med Infect Dis. 2021 Sep 4;6(3):162. doi: 10.3390/tropicalmed6030162. Trop Med Infect Dis. 2021. PMID: 34564546 Free PMC article.
-
Superinfection Exclusion in Mosquitoes and Its Potential as an Arbovirus Control Strategy.Viruses. 2020 Nov 5;12(11):1259. doi: 10.3390/v12111259. Viruses. 2020. PMID: 33167513 Free PMC article. Review.
-
Characterization of Dengue Virus Serotype 2 Cosmopolitan Genotype Circulating in Colombia.Am J Trop Med Hyg. 2023 Oct 9;109(6):1298-1302. doi: 10.4269/ajtmh.23-0375. Print 2023 Dec 6. Am J Trop Med Hyg. 2023. PMID: 37972339 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

