The lncRNA HOTAIR impacts on mesenchymal stem cells via triple helix formation
- PMID: 27634931
- PMCID: PMC5159544
- DOI: 10.1093/nar/gkw802
The lncRNA HOTAIR impacts on mesenchymal stem cells via triple helix formation
Abstract
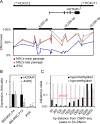
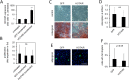
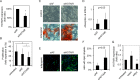
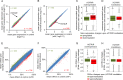
There is a growing perception that long non-coding RNAs (lncRNAs) modulate cellular function. In this study, we analyzed the role of the lncRNA HOTAIR in mesenchymal stem cells (MSCs) with particular focus on senescence-associated changes in gene expression and DNA-methylation (DNAm). HOTAIR binding sites were enriched at genomic regions that become hypermethylated with increasing cell culture passage. Overexpression and knockdown of HOTAIR inhibited or stimulated adipogenic differentiation of MSCs, respectively. Modification of HOTAIR expression evoked only very moderate effects on gene expression, particularly of polycomb group target genes. Furthermore, overexpression and knockdown of HOTAIR resulted in DNAm changes at HOTAIR binding sites. Five potential triple helix forming domains were predicted within the HOTAIR sequence based on reverse Hoogsteen hydrogen bonds. Notably, the predicted triple helix target sites for these HOTAIR domains were also enriched in differentially expressed genes and close to DNAm changes upon modulation of HOTAIR Electrophoretic mobility shift assays provided further evidence that HOTAIR domains form RNA-DNA-DNA triplexes with predicted target sites. Our results demonstrate that HOTAIR impacts on differentiation of MSCs and that it is associated with senescence-associated DNAm. Targeting of epigenetic modifiers to relevant loci in the genome may involve triple helix formation with HOTAIR.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
Senescence-associated DNA methylation is stochastically acquired in subpopulations of mesenchymal stem cells.Aging Cell. 2017 Feb;16(1):183-191. doi: 10.1111/acel.12544. Epub 2016 Oct 26. Aging Cell. 2017. PMID: 27785870 Free PMC article.
-
Detection of RNA-DNA binding sites in long noncoding RNAs.Nucleic Acids Res. 2019 Apr 8;47(6):e32. doi: 10.1093/nar/gkz037. Nucleic Acids Res. 2019. PMID: 30698727 Free PMC article.
-
Long Non-coding RNA-mRNA Correlation Analysis Reveals the Potential Role of HOTAIR in Pathogenesis of Sporadic Thoracic Aortic Aneurysm.Eur J Vasc Endovasc Surg. 2017 Sep;54(3):303-314. doi: 10.1016/j.ejvs.2017.06.010. Epub 2017 Jul 27. Eur J Vasc Endovasc Surg. 2017. PMID: 28757056
-
Epigenetic regulation in human melanoma: past and future.Epigenetics. 2015;10(2):103-21. doi: 10.1080/15592294.2014.1003746. Epigenetics. 2015. PMID: 25587943 Free PMC article. Review.
-
The molecular mechanism of HOTAIR in tumorigenesis, metastasis, and drug resistance.Acta Biochim Biophys Sin (Shanghai). 2014 Dec;46(12):1011-5. doi: 10.1093/abbs/gmu104. Epub 2014 Nov 10. Acta Biochim Biophys Sin (Shanghai). 2014. PMID: 25385164 Review.
Cited by
-
Bioinformatics and Experimental Analyses Reveal Immune-Related LncRNA-mRNA Pair AC011483.1-CCR7 as a Biomarker and Therapeutic Target for Ischemic Cardiomyopathy.Int J Mol Sci. 2022 Oct 9;23(19):11994. doi: 10.3390/ijms231911994. Int J Mol Sci. 2022. PMID: 36233294 Free PMC article.
-
New targeted approaches for epigenetic age predictions.BMC Biol. 2020 Jun 24;18(1):71. doi: 10.1186/s12915-020-00807-2. BMC Biol. 2020. PMID: 32580727 Free PMC article.
-
Senescence-associated DNA methylation is stochastically acquired in subpopulations of mesenchymal stem cells.Aging Cell. 2017 Feb;16(1):183-191. doi: 10.1111/acel.12544. Epub 2016 Oct 26. Aging Cell. 2017. PMID: 27785870 Free PMC article.
-
LncRNA-Smad7 mediates cross-talk between Nodal/TGF-β and BMP signaling to regulate cell fate determination of pluripotent and multipotent cells.Nucleic Acids Res. 2022 Oct 14;50(18):10526-10543. doi: 10.1093/nar/gkac780. Nucleic Acids Res. 2022. PMID: 36134711 Free PMC article.
-
LncRNAs down-regulate Myh1, Casr, and Mis18a expression in the Substantia Nigra of aged male rats.Aging (Albany NY). 2019 Oct 2;11(19):8313-8328. doi: 10.18632/aging.102321. Epub 2019 Oct 2. Aging (Albany NY). 2019. PMID: 31576812 Free PMC article.
References
-
- Dominici M., Le Blanc K., Mueller I., Slaper-Cortenbach I., Marini F., Krause D., Deans R., Keating A., Prockop D., Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. - PubMed
-
- Roobrouck V.D., Ulloa-Montoya F., Verfaillie C.M. Self-renewal and differentiation capacity of young and aged stem cells. Exp. Cell Res. 2008;314:1937–1944. - PubMed
-
- Koch C.M., Wagner W. Epigenetic biomarker to determine replicative senescence of cultured cells. Methods Mol. Biol. 2013;1048:309–321. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

