Virus Matryoshka: A Bacteriophage Particle-Guided Molecular Assembly Approach to a Monodisperse Model of the Immature Human Immunodeficiency Virus
- PMID: 27634413
- PMCID: PMC6810630
- DOI: 10.1002/smll.201601712
Virus Matryoshka: A Bacteriophage Particle-Guided Molecular Assembly Approach to a Monodisperse Model of the Immature Human Immunodeficiency Virus
Abstract
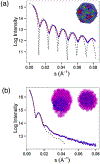
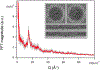
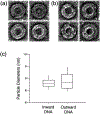
Immature human immunodeficiency virus type 1 (HIV-1) is approximately spherical, but is constructed from a hexagonal lattice of the Gag protein. As a hexagonal lattice is necessarily flat, the local symmetry cannot be maintained throughout the structure. This geometrical frustration presumably results in bending stress. In natural particles, the stress is relieved by incorporation of packing defects, but the magnitude of this stress and its significance for the particles is not known. In order to control this stress, we have now assembled the Gag protein on a quasi-spherical template derived from bacteriophage P22. This template is monodisperse in size and electron-transparent, enabling the use of cryo-electron microscopy in structural studies. These templated assemblies are far less polydisperse than any previously described virus-like particles (and, while constructed according to the same lattice as natural particles, contain almost no packing defects). This system gives us the ability to study the relationship between packing defects, curvature and elastic energy, and thermodynamic stability. As Gag is bound to the P22 template by single-stranded DNA, treatment of the particles with DNase enabled us to determine the intrinsic radius of curvature of a Gag lattice, unconstrained by DNA or a template. We found that this intrinsic radius is far larger than that of a virion or P22-templated particle. We conclude that Gag is under elastic strain in a particle; this has important implications for the kinetics of shell growth, the stability of the shell, and the type of defects it will assume as it grows.
Keywords: HIV; self-assembly; strain; virus shells.
© 2016 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures


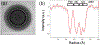
Similar articles
-
Cryo-electron microscopy reveals conserved and divergent features of gag packing in immature particles of Rous sarcoma virus and human immunodeficiency virus.J Mol Biol. 2006 Jan 6;355(1):157-68. doi: 10.1016/j.jmb.2005.10.025. Epub 2005 Nov 2. J Mol Biol. 2006. PMID: 16289202
-
Identification of a Structural Element in HIV-1 Gag Required for Virus Particle Assembly and Maturation.mBio. 2018 Oct 16;9(5):e01567-18. doi: 10.1128/mBio.01567-18. mBio. 2018. PMID: 30327442 Free PMC article.
-
Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle.Curr Biol. 1997 Oct 1;7(10):729-38. doi: 10.1016/s0960-9822(06)00331-9. Curr Biol. 1997. PMID: 9368755
-
Distinct Morphology of Human T-Cell Leukemia Virus Type 1-Like Particles.Viruses. 2016 May 11;8(5):132. doi: 10.3390/v8050132. Viruses. 2016. PMID: 27187442 Free PMC article.
-
Multiple, Switchable Protein:RNA Interactions Regulate Human Immunodeficiency Virus Type 1 Assembly.Annu Rev Virol. 2018 Sep 29;5(1):165-183. doi: 10.1146/annurev-virology-092917-043448. Epub 2018 Jul 26. Annu Rev Virol. 2018. PMID: 30048218 Review.
Cited by
-
Analytical Techniques to Characterize the Structure, Properties, and Assembly of Virus Capsids.Anal Chem. 2019 Jan 2;91(1):622-636. doi: 10.1021/acs.analchem.8b04824. Epub 2018 Dec 3. Anal Chem. 2019. PMID: 30383361 Free PMC article. Review.
-
Bacteriophage P22 Capsid as a Pluripotent Nanotechnology Tool.Viruses. 2023 Feb 13;15(2):516. doi: 10.3390/v15020516. Viruses. 2023. PMID: 36851730 Free PMC article. Review.
-
Defects and Chirality in the Nanoparticle-Directed Assembly of Spherocylindrical Shells of Virus Coat Proteins.ACS Nano. 2018 Jun 26;12(6):5323-5332. doi: 10.1021/acsnano.8b00069. Epub 2018 Apr 25. ACS Nano. 2018. PMID: 29694012 Free PMC article.
-
Beyond icosahedral symmetry in packings of proteins in spherical shells.Proc Natl Acad Sci U S A. 2017 Aug 22;114(34):9014-9019. doi: 10.1073/pnas.1706825114. Epub 2017 Aug 8. Proc Natl Acad Sci U S A. 2017. PMID: 28790186 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

