Segregation between SMCHD1 mutation, D4Z4 hypomethylation and Facio-Scapulo-Humeral Dystrophy: a case report
- PMID: 27634379
- PMCID: PMC5025538
- DOI: 10.1186/s12881-016-0328-9
Segregation between SMCHD1 mutation, D4Z4 hypomethylation and Facio-Scapulo-Humeral Dystrophy: a case report
Abstract
Background: The main form of Facio-Scapulo-Humeral muscular Dystrophy is linked to copy number reduction of the 4q D4Z4 macrosatellite (FSHD1). In 5 % of cases, FSHD phenotype appears in the absence of D4Z4 reduction (FSHD2). In 70-80 % of these patients, variants of the SMCHD1 gene segregate with 4qA haplotypes and D4Z4 hypomethylation.
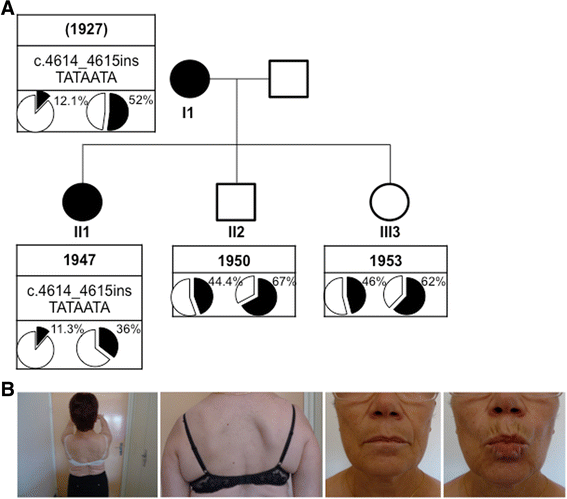
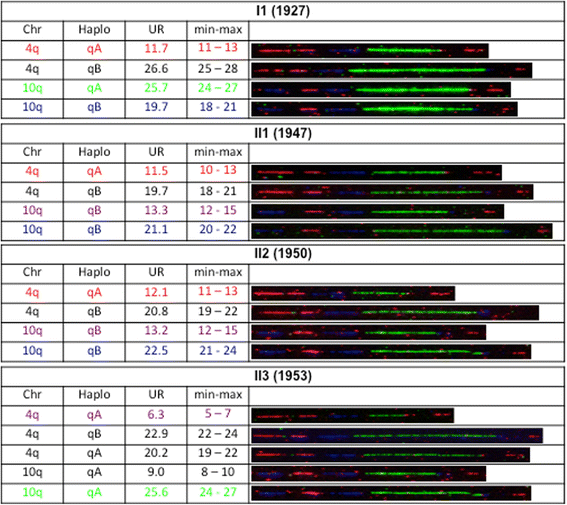
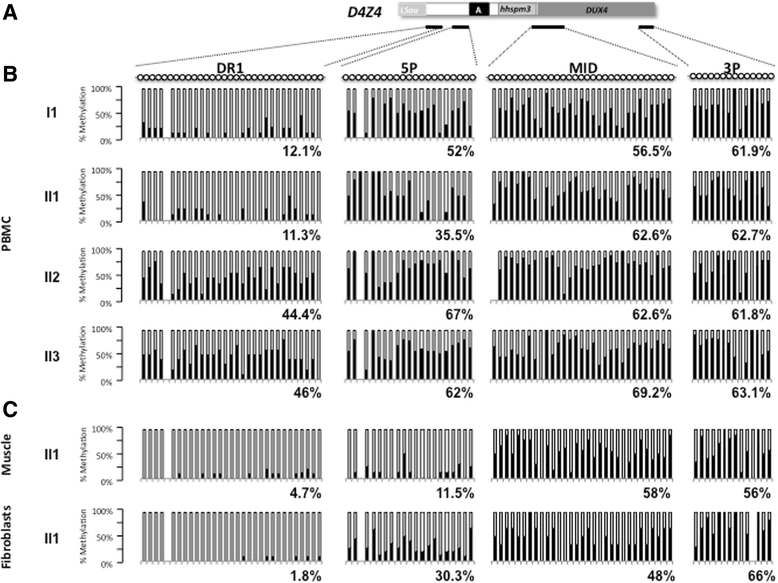
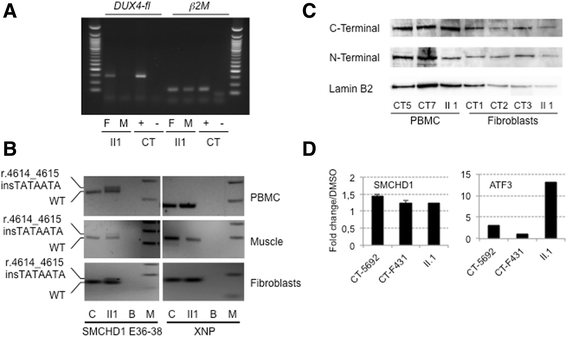
Case presentation: We report a family presenting with neuromuscular symptoms reminiscent of FSHD but without D4Z4 copy reduction. We characterized the 4q35 region using molecular combing, searched for mutation in the SMCHD1 gene and determined D4Z4 methylation level by sodium bisulfite sequencing. We further investigated the impact of the SMCHD1 mutation at the protein level and on the NMD-dependent degradation of transcript. In muscle, we observe moderate but significant reduction in D4Z4 methylation, not correlated with DUX4-fl expression. Exome sequencing revealed a heterozygous insertion of 7 bp in exon 37 of the SMCHD1 gene producing a loss of frame with premature stop codon 4 amino acids after the insertion (c.4614-4615insTATAATA). Both wild-type and mutated transcripts are detected.
Conclusion: The truncated protein is absent and the full-length protein level is similar in patients and controls indicating that in this family, FSHD is not associated with SMCHD1 haploinsufficiency.
Keywords: DNA combing; DNA methylation; DUX4; Facio-Scapulo-Humeral Dystrophy; Haploinsufficiency; SMCHD1.
Figures
Similar articles
-
Double SMCHD1 variants in FSHD2: the synergistic effect of two SMCHD1 variants on D4Z4 hypomethylation and disease penetrance in FSHD2.Eur J Hum Genet. 2016 Jan;24(1):78-85. doi: 10.1038/ejhg.2015.55. Epub 2015 Mar 18. Eur J Hum Genet. 2016. PMID: 25782668 Free PMC article.
-
Diagnostic approach for FSHD revisited: SMCHD1 mutations cause FSHD2 and act as modifiers of disease severity in FSHD1.Eur J Hum Genet. 2015 Jun;23(6):808-16. doi: 10.1038/ejhg.2014.191. Epub 2014 Nov 5. Eur J Hum Genet. 2015. PMID: 25370034 Free PMC article.
-
Genetic and epigenetic characteristics of FSHD-associated 4q and 10q D4Z4 that are distinct from non-4q/10q D4Z4 homologs.Hum Mutat. 2014 Aug;35(8):998-1010. doi: 10.1002/humu.22593. Epub 2014 Jun 24. Hum Mutat. 2014. PMID: 24838473 Free PMC article.
-
Facioscapulohumeral muscular dystrophy.Biochim Biophys Acta. 2015 Apr;1852(4):607-14. doi: 10.1016/j.bbadis.2014.05.021. Epub 2014 May 29. Biochim Biophys Acta. 2015. PMID: 24882751 Review.
-
Facioscapulohumeral Muscular Dystrophy.Compr Physiol. 2017 Sep 12;7(4):1229-1279. doi: 10.1002/cphy.c160039. Compr Physiol. 2017. PMID: 28915324 Review.
Cited by
-
SMCHD1 is involved in de novo methylation of the DUX4-encoding D4Z4 macrosatellite.Nucleic Acids Res. 2019 Apr 8;47(6):2822-2839. doi: 10.1093/nar/gkz005. Nucleic Acids Res. 2019. PMID: 30698748 Free PMC article.
-
Does DNA Methylation Matter in FSHD?Genes (Basel). 2020 Feb 28;11(3):258. doi: 10.3390/genes11030258. Genes (Basel). 2020. PMID: 32121044 Free PMC article. Review.
-
Digenic Inheritance of Shortened Repeat Units of the D4Z4 Region and a Loss-of-Function Variant in SMCHD1 in a Family With FSHD.Front Neurol. 2018 Nov 28;9:1027. doi: 10.3389/fneur.2018.01027. eCollection 2018. Front Neurol. 2018. PMID: 30546343 Free PMC article.
-
SMCHD1 mutation spectrum for facioscapulohumeral muscular dystrophy type 2 (FSHD2) and Bosma arhinia microphthalmia syndrome (BAMS) reveals disease-specific localisation of variants in the ATPase domain.J Med Genet. 2019 Oct;56(10):693-700. doi: 10.1136/jmedgenet-2019-106168. Epub 2019 Jun 26. J Med Genet. 2019. PMID: 31243061 Free PMC article.
-
The variability of SMCHD1 gene in FSHD patients: evidence of new mutations.Hum Mol Genet. 2019 Dec 1;28(23):3912-3920. doi: 10.1093/hmg/ddz239. Hum Mol Genet. 2019. PMID: 31600781 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

