Familial Amyotrophic Lateral Sclerosis-linked Mutations in Profilin 1 Exacerbate TDP-43-induced Degeneration in the Retina of Drosophila melanogaster through an Increase in the Cytoplasmic Localization of TDP-43
- PMID: 27634045
- PMCID: PMC5095402
- DOI: 10.1074/jbc.M116.729152
Familial Amyotrophic Lateral Sclerosis-linked Mutations in Profilin 1 Exacerbate TDP-43-induced Degeneration in the Retina of Drosophila melanogaster through an Increase in the Cytoplasmic Localization of TDP-43
Abstract
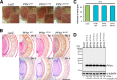
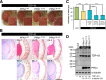
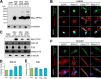
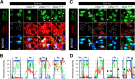
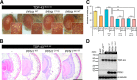
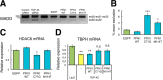
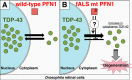
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disease characterized by progressive and selective loss of motor neurons. Causative genes for familial ALS (fALS), e.g. TARDBP or FUS/TLS, have been found, among which mutations within the profilin 1 (PFN1) gene have recently been identified in ALS18. To elucidate the mechanism whereby PFN1 mutations lead to neuronal death, we generated transgenic Drosophila melanogaster overexpressing human PFN1 in the retinal photoreceptor neurons. Overexpression of wild-type or fALS mutant PFN1 caused no degenerative phenotypes in the retina. Double overexpression of fALS mutant PFN1 and human TDP-43 markedly exacerbated the TDP-43-induced retinal degeneration, i.e. vacuolation and thinning of the retina, whereas co-expression of wild-type PFN1 did not aggravate the degenerative phenotype. Notably, co-expression of TDP-43 with fALS mutant PFN1 increased the cytoplasmic localization of TDP-43, the latter remaining in nuclei upon co-expression with wild-type PFN1, whereas co-expression of TDP-43 lacking the nuclear localization signal with the fALS mutant PFN1 did not aggravate the retinal degeneration. Knockdown of endogenous Drosophila PFN1 did not alter the degenerative phenotypes of the retina in flies overexpressing wild-type TDP-43 These data suggest that ALS-linked PFN1 mutations exacerbate TDP-43-induced neurodegeneration in a gain-of-function manner, possibly by shifting the localization of TDP-43 from nuclei to cytoplasm.
Keywords: Drosophila; TAR DNA-binding protein 43 (TDP-43) (TARDBP); amyotrophic lateral sclerosis (ALS) (Lou Gehrig disease); fused in sarcoma (FUS); neurodegeneration; neurodegenerative disease; profilin.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


Similar articles
-
RNA binding mediates neurotoxicity in the transgenic Drosophila model of TDP-43 proteinopathy.Hum Mol Genet. 2013 Nov 15;22(22):4474-84. doi: 10.1093/hmg/ddt296. Epub 2013 Jun 25. Hum Mol Genet. 2013. PMID: 23804749
-
Gain-of-function profilin 1 mutations linked to familial amyotrophic lateral sclerosis cause seed-dependent intracellular TDP-43 aggregation.Hum Mol Genet. 2016 Apr 1;25(7):1420-33. doi: 10.1093/hmg/ddw024. Epub 2016 Jan 28. Hum Mol Genet. 2016. PMID: 26908597
-
Self-assembly of FUS through its low-complexity domain contributes to neurodegeneration.Hum Mol Genet. 2018 Apr 15;27(8):1353-1365. doi: 10.1093/hmg/ddy046. Hum Mol Genet. 2018. PMID: 29425337
-
Molecular basis of amyotrophic lateral sclerosis.Prog Neuropsychopharmacol Biol Psychiatry. 2011 Mar 30;35(2):370-2. doi: 10.1016/j.pnpbp.2010.07.017. Epub 2010 Jul 23. Prog Neuropsychopharmacol Biol Psychiatry. 2011. PMID: 20655970 Review.
-
Amyotrophic Lateral Sclerosis Genes in Drosophila melanogaster.Int J Mol Sci. 2021 Jan 18;22(2):904. doi: 10.3390/ijms22020904. Int J Mol Sci. 2021. PMID: 33477509 Free PMC article. Review.
Cited by
-
The ALS-inducing factors, TDP43A315T and SOD1G93A, directly affect and sensitize sensory neurons to stress.Sci Rep. 2018 Nov 8;8(1):16582. doi: 10.1038/s41598-018-34510-8. Sci Rep. 2018. PMID: 30410094 Free PMC article.
-
A Drosophila model of ALS reveals a partial loss of function of causative human PFN1 mutants.Hum Mol Genet. 2017 Jun 1;26(11):2146-2155. doi: 10.1093/hmg/ddx112. Hum Mol Genet. 2017. PMID: 28379367 Free PMC article.
-
The physiological and pathological biophysics of phase separation and gelation of RNA binding proteins in amyotrophic lateral sclerosis and fronto-temporal lobar degeneration.Brain Res. 2018 Aug 15;1693(Pt A):11-23. doi: 10.1016/j.brainres.2018.04.036. Epub 2018 Apr 30. Brain Res. 2018. PMID: 29723523 Free PMC article. Review.
-
Protein Quality Control and the Amyotrophic Lateral Sclerosis/Frontotemporal Dementia Continuum.Front Mol Neurosci. 2017 May 10;10:119. doi: 10.3389/fnmol.2017.00119. eCollection 2017. Front Mol Neurosci. 2017. PMID: 28539871 Free PMC article.
-
Profilin Directly Promotes Microtubule Growth through Residues Mutated in Amyotrophic Lateral Sclerosis.Curr Biol. 2017 Nov 20;27(22):3535-3543.e4. doi: 10.1016/j.cub.2017.10.002. Epub 2017 Nov 9. Curr Biol. 2017. PMID: 29129529 Free PMC article.
References
-
- Rosen D. R., Siddique T., Patterson D., Figlewicz D. A., Sapp P., Hentati A., Donaldson D., Goto J., O'Regan J. P., Deng H. X., Rahmani Z., Krizus A., McKenna-Yasek D., Cayabyab A., Gaston S. M., et al. (1993) Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362, 59–62 - PubMed
-
- Sreedharan J., Blair I. P., Tripathi V. B., Hu X., Vance C., Rogelj B., Ackerley S., Durnall J. C., Williams K. L., Buratti E., Baralle F., de Belleroche J., Mitchell J. D., Leigh P. N., Al-Chalabi A., et al. (2008) TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 319, 1668–1672 - PMC - PubMed
-
- Yokoseki A., Shiga A., Tan C. F., Tagawa A., Kaneko H., Koyama A., Eguchi H., Tsujino A., Ikeuchi T., Kakita A., Okamoto K., Nishizawa M., Takahashi H., and Onodera O. (2008) TDP-43 mutation in familial amyotrophic lateral sclerosis. Ann. Neurol. 63, 538–542 - PubMed
-
- Kabashi E., Valdmanis P. N., Dion P., Spiegelman D., McConkey B. J., Vande Velde C., Bouchard J. P., Lacomblez L., Pochigaeva K., Salachas F., Pradat P. F., Camu W., Meininger V., Dupre N., and Rouleau G. A. (2008) TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat. Genet. 40, 572–574 - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

