Analysis of long non-coding RNA expression profiles in pancreatic ductal adenocarcinoma
- PMID: 27628540
- PMCID: PMC5024322
- DOI: 10.1038/srep33535
Analysis of long non-coding RNA expression profiles in pancreatic ductal adenocarcinoma
Abstract
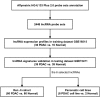
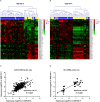
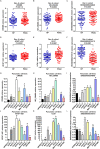
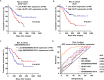
Pancreatic ductal adenocarcinoma (PDAC) remains one of the most aggressive and lethal malignancies. Long non-coding RNAs (lncRNAs) are a novel class of non-protein-coding transcripts that have been implicated in cancer biogenesis and prognosis. By repurposing microarray probes, we herein analysed the lncRNA expression profiles in two public PDAC microarray datasets and identified 34 dysregulated lncRNAs in PDAC. In addition, the expression of 6 selected lncRNAs was confirmed in Ren Ji cohort and pancreatic cell lines, and their association with 80 PDAC patients' clinicopathological features and prognosis was investigated. Results indicated that AFAP1-AS1, UCA1 and ENSG00000218510 might be involved in PDAC progression and significantly associated with overall survival of PDAC. UCA1 and ENSG00000218510 expression status may serve as independent prognostic biomarkers for overall survival of PDAC. Gene set enrichment analysis (GSEA) analysis suggested that high AFAP1-AS1, UCA1 and low ENSG00000218510 expression were correlated with several tumorigenesis related pathways. Functional experiments demonstrated that AFAP1-AS1 and UCA1 were required for efficient invasion and/or proliferation promotion in PDAC cell lines, while ENSG00000218510 acted the opposite. Our findings provide novel information on lncRNAs expression profiles which might be beneficial to the precise diagnosis, subcategorization and ultimately, the individualized therapy of PDAC.
Figures
Similar articles
-
High expression of AFAP1-AS1 is associated with poor survival and short-term recurrence in pancreatic ductal adenocarcinoma.J Transl Med. 2015 Apr 30;13:137. doi: 10.1186/s12967-015-0490-4. J Transl Med. 2015. PMID: 25925763 Free PMC article.
-
An increased expression of long non-coding RNA PANDAR promotes cell proliferation and inhibits cell apoptosis in pancreatic ductal adenocarcinoma.Biomed Pharmacother. 2017 Nov;95:685-691. doi: 10.1016/j.biopha.2017.08.124. Epub 2017 Sep 7. Biomed Pharmacother. 2017. PMID: 28886528
-
Analysis of distinct long noncoding RNA transcriptional fingerprints in pancreatic ductal adenocarcinoma.Cancer Med. 2017 Mar;6(3):673-680. doi: 10.1002/cam4.1027. Epub 2017 Feb 21. Cancer Med. 2017. PMID: 28220683 Free PMC article.
-
The role of long non-coding RNA AFAP1-AS1 in human malignant tumors.Pathol Res Pract. 2018 Oct;214(10):1524-1531. doi: 10.1016/j.prp.2018.08.014. Epub 2018 Aug 20. Pathol Res Pract. 2018. PMID: 30173945 Review.
-
Roles of lncRNAs in pancreatic ductal adenocarcinoma: Diagnosis, treatment, and the development of drug resistance.Hepatobiliary Pancreat Dis Int. 2023 Apr;22(2):128-139. doi: 10.1016/j.hbpd.2022.12.002. Epub 2022 Dec 14. Hepatobiliary Pancreat Dis Int. 2023. PMID: 36543619 Review.
Cited by
-
Overexpression of lncRNA AFAP1-AS1 promotes cell proliferation and invasion in gastric cancer.Oncol Lett. 2019 Sep;18(3):3211-3217. doi: 10.3892/ol.2019.10640. Epub 2019 Jul 22. Oncol Lett. 2019. PMID: 31452798 Free PMC article.
-
Long Non-Coding RNAs in Pancreatic Cancer: Biologic Functions, Mechanisms, and Clinical Significance.Cancers (Basel). 2022 Apr 24;14(9):2115. doi: 10.3390/cancers14092115. Cancers (Basel). 2022. PMID: 35565245 Free PMC article. Review.
-
Microarray Analysis of the Expression Profile of Long Non-Coding RNAs Indicates lncRNA RP11-263F15.1 as a Biomarker for Diagnosis and Prognostic Prediction of Pancreatic Ductal Adenocarcinoma.J Cancer. 2017 Aug 22;8(14):2740-2755. doi: 10.7150/jca.18073. eCollection 2017. J Cancer. 2017. PMID: 28928863 Free PMC article.
-
Meta-analysis of the prognostic value of long non-coding RNA AFAP1-AS1 for cancer patients in China.Oncotarget. 2017 Dec 21;9(8):8100-8110. doi: 10.18632/oncotarget.23568. eCollection 2018 Jan 30. Oncotarget. 2017. PMID: 29487718 Free PMC article.
-
Revealing the potential role of hub metabolism-related genes and their correlation with immune cells in acute ischemic stroke.IET Syst Biol. 2024 Aug;18(4):129-142. doi: 10.1049/syb2.12095. Epub 2024 Jun 8. IET Syst Biol. 2024. PMID: 38850201 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

